Article Text
Abstract
This review is focused on the challenges in standardising and optimising molecular testing workflow in cytopathology. Although cytological samples yield optimal quality DNA, whose minimal amounts in most cases suffice even for multigene mutational profiling, the success of molecular testing is strongly dependent on standardised preanalytical protocols for maximising DNA yield and quality. Sample cytopreparation influences, even more, the quality of RNA and consequently the potential success of reverse transcription-PCR. Here, the educational and technical involvement of the cytopathologist as a relevant component of a multidisciplinary team, in the issues related to test request, specimen collection, fixation, processing, staining, tumour fraction enrichment, DNA quality/quantity assessment and storage conditions is discussed. In addition, the specific sample requirements related to more recent technological developments are examined, underlining the modern role of the cytopathologist, whose continuous education is crucial to meet the opportunities of molecular medicine.
- personalized medicine
- cancer
- cytopathology
- fine-needle aspiration
- molecular cytopathology
- polymerase chain reaction
- next generation sequencing
- mutational analysis
- preanalytic
- direct smear
- cell block
Statistics from Altmetric.com
- personalized medicine
- cancer
- cytopathology
- fine-needle aspiration
- molecular cytopathology
- polymerase chain reaction
- next generation sequencing
- mutational analysis
- preanalytic
- direct smear
- cell block
Introduction
Molecular cytopathology, a rapidly evolving field of modern cytopathology, features an increasing number, variety, breadth and depth of tests, which underlines the effective interplay between genomics and cytology.1 Challenging cases classified as atypical or as of undetermined significance may be further stratified into high- and low-risk groups by the demonstration of specific oncogenic mutations.2 Moreover, by the development of personalised/precision medicine, cancer gene testing on cytological samples from patients with surgically unresectable, high-stage, locally advanced, recurrent or metastatic malignancies is crucial.3 Although fine needle aspiration (FNA) biopsy, a rapid, efficient and minimally invasive technique, and core needle biopsy (CNB) represent complementary methods to sample superficial and deep-seated lesions, the use of FNA for gene testing is advantageous over CNB in several respects. Despite a wide range of cytopreparations, fixation and staining techniques, usually, most of FNA have higher tumour fraction, ensure a wider sampling of the targeted lesion and offer a better quality DNA and an effective triage for ancillary studies when coupled with rapid onsite evaluation (ROSE).2 More recently, cytological specimens have also been validated for next-generation sequencing (NGS) to simultaneously screen different types of mutations in multiple genes and in multiple patient samples using small amounts of input material.4
Cytopathologist’s role
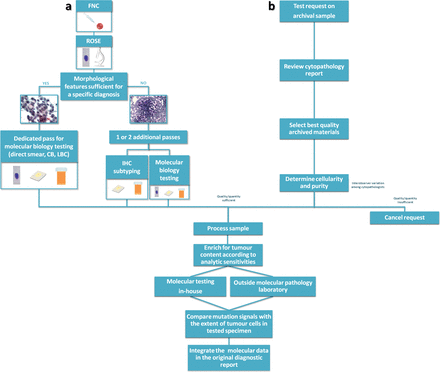
The cytopathologist plays a central role in the workflow of the molecular analysis performed on cytological samples. His/her range of activities and the extent of interactions with cytotechnicians and molecular biologists greatly differ among institutions reflecting local issues and resources. As an example, in our institution, the cytopathologist is the one mainly responsible for those multiple actions cumulatively referred to as preanalytical processing (figure 1). In fact, the cytopathologist has to review cytopathology reports and archived materials to select best quality smears or representative cell block (CB) sections to determine the cellularity and purity of the tumour sample being submitted for biomarker testing, having the responsibility to cancel the request for molecular assay whenever the cellularity is below the analytical sensitivity of the molecular assay. In other institutions, the adequacy assessment is best made by the external molecular laboratory, which should provide a written report explaining the reason of test cancellation.
Cytopathologist role in molecular testing. The cytopathologist plays a different role when molecular testing is performed prospectively at the moment of the diagnosis (A) or when the test is ordered by the oncologist on an archival sample (B). In (A), the cytopathologist is responsible also for the triage decision on how to manage the specimen. In this setting, ROSE is crucial to ensure sample adequacy for morphological diagnosis, ancillary techniques and molecular analysis. In (B), the cytopathologist has to review cytopathology reports and archived materials to select the best quality sample among several preparation types with varying suitability, having the responsibility to cancel the request for molecular assay whenever the cellularity, even after tumour cell enrichment, is below the analytical sensitivity of the molecular assay. Regardless of the test being performed in-house or in referral laboratories, the cytopathologist needs to evaluate critically the results before integration in the original cytological diagnostic report. CB, cell block; FNC, fine needle cytology; IHC, immunohistochemistry; LBC, liquid-based cytology; ROSE, rapid onsite evaluation.
Similar to surgical pathologists, there is a wide interobserver variation also among cytopathologists in estimating tumour fraction, and even in the same institution, cancellation rates vary widely among cytopathologists.5 Care should be taken to identify viable tumour areas in which the tumour ratio is optimal, and the percentage of inflammatory cells and potentially amplification inhibitors (such as mucin, melanin and tumour cell necrosis) is minimal.2 Since various mutational assays have different analytical sensitivities, the cytopathologist (or the technician) should enrich for tumour content to a level that is acceptable for the assay being used. Once the results of the genotyping analysis are received, the cytopathologist needs knowledge of the molecular diagnosis and of available treatment strategies, taking care to integrate the molecular data in its original diagnostic report.
Test request
As a general rule, the test request should be made appropriately to ensure that every patient who needs a test is offered one in a timely manner, while avoiding unnecessary procedures. The test is usually requested by the oncologists and less frequently by other specialists, including surgeons and interventional radiologists. Ideally, rather than by a single specialist, test request should be made multidisciplinary (tumour board).6 Also, in light of the increased awareness among patients and their families of the novel technological and therapeutic opportunities, the tumour board should ensure that the needs of a precise cytological diagnosis and of multiple predictive assays would simultaneously be met.6 Thus, the effective communication among the laboratory, the oncologists and the cytopathologist is crucial to plan effective sampling strategies to ensure that adequate tissue amount is obtained.7 As a matter of the fact, the cytopathologist may not know whether the patient is a candidate for surgery or for targeted therapy. Thus, the cytological sample is not the optimal testing approach when a larger resection specimen is subsequently available for analysis.7 Similarly, for diagnoses made on a metastatic or recurrent lesion, the cytopathologist should be informed whether any prior specimen of the same patient has already been tested. Previous chemotherapy regimens can change gene expression and mutation status and should be documented on the request form. In some cases, patients with poor performance status may still be considered candidates for testing, as clinical response without significant side effects may follow the detection of a targetable genomic alteration.7
Rather than on oncologist’s demand, the automatic (reflex) testing by cytopathologists, based on diagnosis and tissue availability, can be more efficient. Reflex testing avoids the costs in time and money of specimen retrieval from pathology archives and the treatment delay for patients who are found to harbour a targetable molecular alteration.8 However, molecular testing is expensive, and as molecular biomarkers are evolving rapidly over time, new targets may be identified in the interval between diagnosis and recurrence.
Reference laboratories
While most academic medical centres are equipped to run molecular testing in-house, the vast majority of pathology services are provided through external molecular laboratories.9 Processes should be established to ensure that specimens with a final cytopathology diagnosis are sent to external molecular pathology laboratories within 3 working days of receiving requests. However, in our practice, the mean time between the oncologist’s request and specimen dispatching is nearly double the recommended time (5.8 working days).9 Noteworthy, delivery times are longer for larger volume pathology departments than for smaller laboratories. Budget, the availability of technical personal and reimbursement issues may be factors influencing these differences. However, once the cytopathologists are made aware of the delays in the processing of the request and the shipping of the tumour samples, corrective actions can be obtained.10 The breakage of slides during transport is also a potential issue; nonetheless, careful packing of slides can prevent this to be a serious drawback to the use of smears.
When determining the centre to select for outside molecular testing, the cytopathologist should ensure that the laboratory is accredited either at the national or at the international level.7 The laboratory should join external quality assurance assessment once or twice a year; however, as only histological samples are usually included in the proficiency testing schemes, the assessment of the quality of testing on cytology remains problematic. It is also relevant that the laboratory staff would include anatomical pathology certified pathologists who verify specimen quantity and quality and supervise specimen selection, interface with clinicians and troubleshoot problems.7
The cytopathologist should be aware of the minimum tumour percentage required by the reference laboratory to accept a specimen. While the analytical sensitivity dictates the burden of tumour that must be present in the tested sample, it is also necessary that the method used has sufficient reference range for a wide spectrum of mutations. The cytopathologist should know whether the reference laboratory routinely microdissects samples and the method of microdissection used. Another key issue to consider is the cytopreparation types validated and accepted for testing.7 Most laboratories will accept cytology CBs, while an additional option that has already reached widespread adoption is the use of direct smears.
Cell block
Formalin-fixed, paraffin-embedded (FFPE) CBs were first introduced in the early 1970s, as an aid to microscopic diagnosis, by highlighting tumour architectural organisation not readily appreciated on other cytological preparations.11 Later, CBs have traditionally been employed by cytopathological laboratories to perform ancillary immunocytochemical stainings. In the era of precision medicine, they represent a useful banked tissue archive for molecular testing, given the regulatory requirement for archival slide retention and the concern to use direct smears.2 Moreover, as most molecular testing clinical practice guideline panels include expert molecular histopathologists, the use of CB sections is usually recommended over smears.2 CBs are usually similar to the traditional histological blocks and, in most cases, do not require additional molecular assay validation12; however, there are multiple CB preparation techniques, such as those based on agar or fibrin, which may require an additional validation based on individual laboratory preferences. Unfortunately, CBs have nucleic acid preservation issues similar to those of histological FFPE. In fact, neutral buffered formalin, the fixative most commonly used for tissue preservation, induces the methylene bridging of bases and the formation of crosslinks between nucleic acids and available proteins and random polymerase errors in nucleotide incorporation, usually being C-T or G-A transitions13; these sequencing artefacts mainly occur when the amounts of template DNA are low, as in the case of DNA obtained from microdissected sections, and DNA treatment with Escherichia coli uracil N-glycosylase before amplification and genotyping on shorter amplicons may be a way to avoid artifactual mutations.13 As far as RNA extraction is concerned, relatively harsh conditions with the inclusion of proteinase K digestion followed by heating steps are employed in an effort to break the methylene bridges. Even with optimised digestion and heating steps, however, it is not possible to completely remove all chemical modifications such as residual methyl groups from FFPE-extracted RNA.14
Cellularity is evaluated by examining an H&E-stained section prepared from the CB; thus, the percentage of tumour cells in deeper sections of the CB used for molecular testing is assessed in an extrapolative fashion inferred but not actually known. When the tumour cellularity is high and more than sufficient for testing, paraffin scrolls can simply be placed directly into a tube for extraction without microdissection, and cellularity assessment of an H&E section taken after the scrolls (postcurl section) may be unnecessary.15 More often, however, CBs feature a low tumour content,16 and in a recent electronic survey among the members of the American Society of Cytopathology and other pathologists, many laboratories shared dissatisfaction with their CB preparation methods.17 As mentioned above, across institutions, there is extreme variability in CB preparation techniques and lack of uniformity with some practices including additional dedicated passes for cellular enrichment.17 Thus, with CBs with low tumour content, unstained sections should be lined up with a corresponding tumour-mapped, H&E-stained slide, with circled tumour-rich areas as a guide for macrodissection or microdissection. Noteworthy, the standard 4–5 µm CB sections do not represent the entire nuclei from the cell and are likely to have lower nucleic acid yields for molecular testing per cell than the whole cells obtained from other non-formalin-fixed cytological substrates.15 Cutting extra, unstained CB sections up front to avoid refacing block would be ideal to save as much tumour tissue as possible for molecular testing, avoiding that ancillary studies are performed in a piecemeal fashion.18
A main disadvantage of using CBs is the inability to assess cellularity and adequacy at the time of procedure because processing is not usually complete until the following day (table 1). Thus, the adequacy assessment of CB preparations is largely based on the ROSE performed on the corresponding direct smears, which may or may not be entirely representative of the CB cellularity.16 17 In fact, usually, the CB represents a pooled specimen from multiple passes, and therefore, the tumour cell population from high-yield needle passes may be eventually diluted by background benign elements from off-target needle passes. This is problematic because the analytical sensitivity of molecular diagnostic assays depends on the percentage of tumour cellularity threshold, below which false-negative results will occur (ie, contaminating benign tissue will be negative for the molecular abnormality being tested).
Advantages and disadvantages of different cytological preparations for mutation testing
Direct smears
Unlike CBs, the additional rigorous validation for each individual molecular assay performed on smear preparations for clinical reporting poses the biggest challenge in using these specimen preparations for ancillary studies.16 However, when a CB is not available, the smears used for diagnosis are the only source of tumour cells testable for molecular studies. As far as manual microdissection is concerned, the direct smears are typically superior to the CBs because the smeared sample is more dispersed with a greater variation in the proportion of tumour/benign in different areas of the slide (table 1). Therefore, it is easier to find and delineate areas of tumour enrichment on smears, even in cases with overall low tumour fraction.16
Among many possible approaches, the usual methods of cell isolation are either via the scraping of the smear or by cell lifting.19 The first procedure is usually carried out by a flat, single-edge scalpel blade to collect all material into a small clump, which is pushed to a corner of the slide. The corner of the slide is placed over the open end of an Eppendorf collecting tube, and the scraped tissue is gently pushed into the tube with the tip of the scalpel blade or a pipette tip. The tissue selection by cell lifting exploits the Pinpoint solution of the Pinpoint Slide DNA Isolation System (Zymo Research) that is applied over the selected area.20 The quantity of Pinpoint solution required is based on the dimension of the tissue area and is calculated according to the manufacturer’s instructions. The solution is spread evenly over the area of interest with the side of a pipette tip and was allowed to air-dry for approximately 30–45 min. After the solution is completely dry and had formed a thin blue film, the embedded tissue together with the dried film is loosened, with a razor blade used to cut around the edge of the film. The film is then peeled from the slide, transferred to an Eppendorf tube and centrifuged briefly so that the tissue could be collected at the bottom of the tube.19
More recently, molecular testing has been validated on DNA extracted from cancer cells isolated from routine smears by cell transfer technique.21 22 This method had already been employed to enable immunohistochemical stainings, and it is based on the use of a special media (Mount Quick) commercially available (Daido Sangyo, Tokyo, Japan). The latter is spread uniformly over the top of the cellular material on de-coverslipped smears. After slide heating, the media is hardened, cut and placed in an Eppendorf tube for DNA extraction and molecular testing.21 This method has several advantages, being inexpensive, easily performed by a technician of the molecular laboratory staff and enabling multiple analysis in selected slide areas, which can be useful when dealing with different cancer cell population components.22
Both alcohol-fixed and air-dried smears are generally suitable for the readily isolation of reasonably stable high-quality DNA and a sound choice for long-term DNA storage, although heat and humidity are potential problematic issues.23 Non-cross-linking alcoholic reagents may yield superior results as RNA fixatives in comparison with aldehydes because they cause little chemical change and usually provide higher quality nucleic acids for molecular testing than do FFPE sections.14 Several studies using previously stained cytology smears have shown that molecular testing can be performed successfully using both Diff-Quik as well as Papanicolaou-stained slides. However, a recent study by Killian et al 24 suggests that Diff-Quik-stained smears should be preferred to Papanicolaou-stained slides. While the latter featured DNA degradation as a function of age, the Diff-Quik-stained smears provided high-quality DNA even if archived for a prolonged period, allowing for the performance of sophisticated molecular diagnostic studies such as high-resolution comparative genomic hybridisation assays.24 Conversely, even more recently, in a cell line based study, Papanicolaou-stained smears yielded optimal DNA quantityand fragment length.25 Interestingly, several studies showed that Diff-Quik smears are as good as CBs and Papanicolaou for NGS testing without significant technical differences.
Although the process of removing the coverslip of archival smears does not compromise the quality of the DNA isolated for molecular studies, it is time consuming.25 To avoid any delay, ROSE, at the time of the FNA procedure, enables the best triage of the sample for diagnosis and ancillary studies, and the selection of a representative slide is maintained uncoverslipped for immediate DNA extraction.20 Alternatively, da Cunha Santos et al proposed the ‘freezer method’.26 Once the slide is frozen, a blade is used to lift off the coverslip, and after xylene soaking, the slide can be then sent for manual microdissection. This method is very fast and could be an important tool for molecular analysis performed on cytology smears.27
When most of the diagnostic cells are on a single slide, the molecular testing will destroy the evidence of tumour cells, which might have medico-legal consequences. To mitigate the medico-legal constraints, smears can be digitally scanned, to record the cytomorphology of representative diagnostic microscopic fields for the archives. In our practice, we experienced that, to digitally record the three-dimensional groups of variable thickness frequently observed in traditional smears, the use of a z-axis scanner is required, which unlike the monoplanar whole slide imaging of histology slides, is time consuming and results in relatively large digital image files.5
Liquid-based cytology
ROSE for tumour cell adequacy is crucial to ensure that the obtained material is sufficient and properly preserved for the identification of malignancy and for biomarker testing (figure 1).2 This can be successfully performed either by a cytopathologist or by a properly trained cytotechnologist or a dedicated clinician.28 Unfortunately, due to budget and staff limitation, ROSE is not always feasible.29 In this setting, liquid-based cytology (LBC) represents a valid alternative to traditional cytology, avoiding the possibility that untrained clinicians may improperly smear and triage the aspirated material, thus limiting artefacts.30
The specimen is simply expelled in its entirety into an alcohol-based fixative, such as CytoLyt (Hologic, Bedford, Massachusetts, USA) or CytoRich Red (Fisher Scientific UK, Loughborough, Leicestershire, England) solutions, and by proprietary instruments, a cell monolayer slide is prepared.30 In our experience, although direct smears show a higher DNA yield and are more cell rich than LBC slides, the differences in adequacy and in mutant rate between the two samples are minimal.30 This may probably reflect the similar effect of methanol-based CytoLyt and ethanol-based smear fixation on DNA preservation (table 1). Conversely, LBC samples fixed with CytoRich Red have shown poorer DNA preservation due to the presence of a small amount (<1%) of formaldehyde that may cause DNA degradation and modification by the cross-linking of cytosine residues on either strands.16 In addition, residual material from CytoLyt samples has been shown to feature optimal RNA integrity being suitable for nucleic acid isolation and subsequent analysis by reverse transcription-PCR (RT-PCR).31 However, RNA degradation was reported in specimens stored for 12 months at room temperature, and long-term storage requires –80°C.14 Interestingly, also, exfoliative oral cytology specimen has shown an adequate RNA preservation.31
Several studies have described using LBC specimens for molecular analysis, either by scraping off cells from the LBC slides or by extracting DNA directly from the rinse solution.32 33 In a recent survey, we reported that the referring cytopathologists more frequently outsource LBC slides rather than vials to referral laboratories; in fact, LBC phial dispatching is impractical as the vials are stored only for a limited period of time with limited long-term DNA stability, and often, the residual solution is not sufficient for testing.30 In addition, the possibility of directly visualising neoplastic cells is preferable, also when comparing the mutation signals with the extent of tumour cells in the tested slide.33 When the low-sensitive direct sequencing method is employed, neoplastic cell enrichment is mandatory12; however, manual microdissection on LBC slides is difficult. In fact, similar to CBs, neoplastic and non-neoplastic components from different in- and off-target FNA passes are pooled together and homogeneously distributed during processing. Laser capture microdissection is expensive and time consuming and hardly feasible in routine clinical setting.32 Alternatively, highly sensitive molecular techniques, such as real-time PCR methods, can be used directly on the DNA extracted from the preserving solution of the phial, without slide preparation.30
Fresh cells
Fresh, unfixed cells may be processed for immediate nucleic acid extraction with excellent results.2 The advantage of a short acquisition time for molecular processing is mostly required to ensure high-integrity RNA for some molecular applications such as complementary DNA labelling for microarray analysis and transcriptome analysis.14 In contrast, RT-PCR or quantitative RT-PCR analysis for fusion gene detection is more tolerant of partially degraded RNA because the design can be based on an analysis of smaller regions of RNA.14 For long-term storage, aliquots can be frozen at 80°C RNA later or similar RNA stabilising solutions and stored in freezers34; fresh cells can also be stored at room temperature for months in Whatman filter paper cards (GE Healthcare Life Sciences, Buckinghamshire, England). The latter method is an easy, fast, inexpensive, operative-friendly procedure and ensures high quality of nucleic acid for molecular testing, but the amount of genetic material that can be extracted from the Flinders Technology Associates (FTA) cards is limited.35 Moreover, the disadvantage of using fresh/frozen/FTA-collected cells is the lack of direct microscopic examination of the tissue specimen from which the DNA/RNA is isolated and false negatives can commonly occur if the sample tested does not have an adequate tumour fraction.23
Nucleic acid sample quantity and quality assessment
The accurate analysis of input nucleic acid sample quantity, purity and integrity is crucial, especially on scant routine cytological samples. Several techniques should be used in a complementary manner, as none of them can provide all the information required to fully characterise the DNA/RNA sourced from a cytological sample.34–36
Spectrophotometer analysis, based on ultraviolet light absorption of a diluted nucleic acid samples read at 260 nm and 280 nm, is largely used to quantify DNA or RNA. Between nucleic acid correlation and absorbance (A), there is a linear correlation able to predict the DNA or RNA quantity in the solution. Pure RNA has an A260/A280 of 2.1, whereas pure DNA will have an A260/A280 of 1.8. Currently, miniaturised automatic platforms, such as NanoDrop (ThermoFisher) spectrophotometer, also allow accurate analyses of small sample sizes.36 It is critical to observe that the optical density reading is a measure of absorption and provide a measure of quantity and not quality or sample integrity.
Fluorimetric assays represent an alternative to spectrophotometric methods.36 The binding of fluorescent dyes to nucleic acids measures the subsequent changes in fluorescence levels. With respect to spectrophotometry, fluorescence-based quantification is more sensitive and precise and may be specific for the nucleic acid of interest. The fluorescence is assessed, by calibrating the sample against a reference specimen, with a known nucleic acid concentration, generating a curve to ensure the linearity of the assay in the range evaluated. As an example, the automated QuBit 2.0 fluorometer (Life Technologies) can be employed to measure a wide range of different nucleic acid concentrations, enabling the accurate analysis of small samples.
Nucleic acid samples can be analysed and compared using instrumentation such as the Agilent 2100 BioAnalyzer, Bio-Rad Experion or the last developed Agilent Tape Station 4200,37 combining capillary electrophoresis with fluorescence by a lab-on-a-chip approach. In fact, the electrophoresis is miniaturised on a chip, featuring glass microchannels and interconnected networks, thus limiting analysis time and input sample requirement. The chip (or the cartridges for Tape Station 4200 only) accommodates wells for samples, gel and an external standard (fragment size ladder).37 The informatic suites that support these instruments are used to determine a relative integrity number for the DNA (only Tape Station 4200) and RNA samples. While the quantitation of nucleic acid input by simple measurement of the DNA concentration in the sample does not provide information on the efficiency of DNA amplification, quantitative PCR assays can be used for quality control for inhibitor checks and to determine the extent of amplifiable DNA.36
DNA input
Reliable, consistent, robust and accurate results from molecular tests using cytological samples will depend on standardised protocols for maximising DNA yield and quality, and preanalytical variables will have a direct impact on the analysis.16
Although very small quantities of input material can be successfully amplified for molecular evaluation thanks to the exponential increase of target DNA via PCR, the yield of DNA is a critical preanalytical factor that determines the success of molecular analysis. Thus, assay validation with low-input DNA levels is crucial to reliably process cytological samples.2 7 In fact, a test that confidently detects a mutation with a specified quantity of input DNA relies on the fact that the tested DNA contains a minimum burden of the mutation. As the input quantity is decreased, the total mutation burden may drop below the lower threshold of detection for the assay. Whenever the tested specimens that do not meet the validated input requirement have a negative result, a disclaimer in the molecular pathology report is needed to indicate that the analytical sensitivity of the assay may be compromised by reduced nucleic acid input.7
A number of variables associated with cytological samples, including the type of, fixative, slide, mounting medium and the tissue extraction methodology, can affect the yield and quality of DNA. A detailed study carried out on cell lines by Dejmek et al 25 reported that spray or ethanol-fixed slides provide better results in terms of DNA quality and yield over air-dried slides.25 Slide type can also have an effect on the DNA yield.19 Clinical laboratories may use a variety of glass slides for the routine processing of smears from aspirates. Fully frosted (FF) slides are useful in low-cellularity aspiration samples, since their high cellular adhesion capability prevents cell loss during fixation in alcohol-based fixative solutions. Conversely, non-frosted (NF) slides have no specialised surface or coating to enhance cellular adhesion, being used when adequate cellularity is not an issue. The positively charged (PC) slides have a specialised surface that electrostatically enhances the adhesion of cellular material. A recent study reported a lower DNA yield for FF slides in comparison with NF and PC, which likely reflects the difficulty in dislodging cells from the crevices of FF slides.19 Thus, although FF slides show better cell retention than other slides, they are more difficult to use for tissue extraction and are not optimal for the DNA yield. On the other side, further investigation is needed to assess the risk of nucleic acid cross-sample contamination, as a result of cell detachment from low-adhesion slides.
Similarly, when cells are directly scraped off from previously stained archival slides by dissection, the DNA yield is higher than that obtained by cell lifting employing the Pinpoint solution.19 Noteworthy, a significantly higher DNA yield was obtained with slides mounted with the low-hazard, organic, polymer-based mounting medium EcoMount (BioCare Medical LLC, Concord, California, USA) when compared with the xylene-based mounting medium Pertex (CellPath, Newtown, Powys, UK).25
Future directions
As technology is advancing at rapid pace, a range of novel techniques is emerging. In particular, NGS and fully automated platforms may necessitate specific sample requirements and dedication from cytopathologist to develop special cytopreparation protocols.
In particular, establishing the minimum number of cells needed to allow an NGS approach from cytology sample is a crucial point. The studies that applied NGS to cytological material had usually a retrospective design, and only samples that featured at least 20% of neoplastic cells were selected, which may not fully reflect current practice. In any case, sample requirement depends on target capture, gene panel and platform types. Illumina NGS usually requires more cells and/or higher DNA input than Ion Torrent NGS; thus, the latter seems to be more efficient with the cytopathologist specimens.5
An example of direct smear from a thyroid FNA, successfully processed by NGS, is reported in figure 2. Even more recently, it was shown that lowering the input DNA concentration below the manufacturer’s recommended threshold of 10 ng (>0.8 ng/µL) is feasible leading to a marked increase in the NGS success rate from 58.6% to 89.8%.5 38
Next-generation sequencing (NGS) on a direct smear. (A) Thyroid fine needle aspiration (FNA) diagnosed as malignant, papillary thyroid cancer. Note the high cellularity (direct smears, Diff-Quik staining, 20× magnification). (B) Genomic variant (BRAFV600E) identified by the Variant Caller software. (C) The BRAFV600E mutation was orthogonally confirmed by real-time PCR.
More relevant than DNA input is the percentage of neoplastic cells; in a low cancer cell background, the preferential amplification of a small number of DNA molecules may be representative only of the benign component, leading to a false-negative result. As a matter of the fact, most NGS assays have a lower limit of mutation detection of 10%, which requires at least 20% of neoplastic cells.39
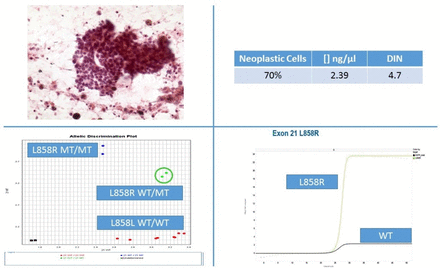
Besides NGS technological improvements, automated allele-specific real-time PCR technology is also advancing at a rapid pace. In particular, the fully automated molecular diagnostic system Idylla (Biocartis, Mechelen, Belgium) is a fascinating technology.40 41 Sample preparation is combined with PCR thermocycling and fluorescence detection of target sequences. Without needing highly skilled staff, within approximately 90 min, the European Community (CE)-in vitro diagnostic use marked Idylla mutational tests can genotype relevant biomarkers. Although the Idylla tests were designed for use with FFPE sections, the Idylla system can also process DNA preparations from cytological samples.42 43 To this end, only few quantity of archival DNA (10 ng), directly pipetted into the cartridge, is sufficient to obtain results in most samples (figure 3).44 45 Conversely, further technological refinements are needed to process scraped cells and to better adapt the automated extraction modalities to stained cytological material.
Archival DNA was extracted from a manual macrodissected Papanicolaou-stained smear of lung adenocarcinoma featuring 70% of neoplastic cells and a concentration of 3.39 ng/µL with a DNA integrity number (DIN) of 4.7. Representative graphs of standard real-time PCR and Idylla are reported both showing L858R epidermal growth factor receptor mutation.
In conclusion, the cytopathologist provides the specimen for molecular diagnostics, among several preparation types with varying suitability, and establishes when, where and how biomarker testing should be performed.9 To this end, modern cytotechnologists and cytopathologists should be dedicated to optimising and standardising cytological sample preparation methods for cytomorphology and for preservation of biomolecular integrity.3
Take home messages
The success of molecular testing on cytology is strongly dependent on standardised preanalytical protocols.
Besides specific analytical issues, dependent on the given molecular technique, appropriate test request, specimen collection, fixation, processing, staining, tumour fraction enrichment, DNA quality/quantity assessment and storage conditions are also crucial.
References
Footnotes
Handling editor Runjan Chetty.
Contributors CB and GT conceived the review and wrote the paper; CB, UM, EV, PP, GV and GT collected and assembled the literature data.
Competing interests None declared.
Patient consent Obtained.
Provenance and peer review Not commissioned; externally peer reviewed.