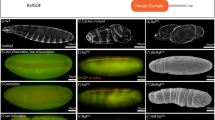
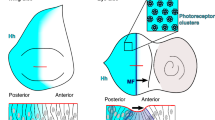
Abstract
Hedgehog (Hh) proteins act through both short-range and long-range signalling to pattern tissues during invertebrate and vertebrate development1. The mechanisms allowing Hedgehog to diffuse over a long distance and to exert its long-range effects are not understood. Here we identify a new Drosophila gene, named tout-velu, that is required for diffusion of Hedgehog. Characterization of tout-velu shows that it encodes an integral membrane protein that belongs to the EXT gene family. Members of thisfamily are involved in the human multiple exostoses syndrome, which affects bone morphogenesis2,3,4. Our results, together with the previous characterization of the role of IndianHedgehog in bone morphogenesis5,6,7, lead us to propose that themultiple exostoses syndrome is associated with abnormal diffusion of Hedgehog proteins. These results show the existence of a new conserved mechanism required for diffusion of Hedgehog.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Johnson, R. L. & Tabin, C. The long and short of Hedgehog signaling. Cell 81, 313–316 (1995).
Ahn, J. et al. Cloning of the putative tumour suppressor gene for hereditary multiple exostoses (EXT1). Nature Genet. 11, 137–143 (1995).
Stickens, D. & Evans, G. A. Isolation and characterization of the murine homolog of the human EXT2 multiple exostoses gene. Biochem. Mol. Med. 61, 16–21 (1997).
Wicklund, C. L., Pauli, R. M., Johnston, D. & Hech, J. T. Natural history study of hereditary multiple exostoses. Am. J. Med. Genet. 55, 43–46 (1995).
Lanske, B. et al. PTH/PTHrP receptor in early development and Indian Hedgehog-regulated bone growth. Science 273, 663–666 (1996).
Vortkamp, A. et al. Regulation of rate of cartilage differentiation by Indian Hedgehog and PTH-related protein. Science 273, 613–622 (1996).
Vortkamp, A. et al. Recapitulation of signals regulating embryonic bone formation during postnatal growth and in fracture repair. Mech. Dev. 71, 65–76 (1998).
Perrimon, N., Lanjuin, A., Arnold, C. & Noll, E. Zygotic lethal mutations with maternal effect phenotypes in Drosophila melanogaster. II. Loci on the second and third chromosomes identified by P-element-induced mutations. Genetics 144, 1681–1692 (1996).
Lee, J. J., von Kessler, D. P., Parks, S. & Beachy, P. A. Secretion and localized transcription suggest a role in positional signaling for products of the segmentation gene hedgehog. Cell 71, 33–50 (1992).
Tabata, T. & Kornberg, T. B. Hedgehog is a signaling protein with a key role in patterning Drosophila imaginal discs. Cell 76, 89–102 (1994).
Basler, K. & Struhl, G. Compartment boundaries and the control of Drosophila limb pattern by Hedgehog protein. Nature 368, 208–214 (1994).
Chen, Y. & Struhl, G. Dual roles for patched in sequestering and transducing Hedgehog. Cell 87, 553–563 (1996).
Alexandre, C., Jacinto, A. & Ingham, P. W. Transcriptional activation of Hedgehog target genes in Drosophila is mediated directly by the Cubitus interruptus protein, a member of the GLI family of zinc finger DNA-binding proteins. Genes Dev. 10, 2003–2013 (1996).
Strigini, M. & Cohen, S. M. AHedgehog activity gradient contributes to AP axial patterning of the Drosophila wing. Development 124, 4697–4705 (1997).
Aza-Blanc, P., Ramirez-Weber, F. A., Laget, M. P., Schwartz, C. & Kornberg, T. B. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell 89, 1043–1053 (1997).
Riemer, D. et al. Expression of Drosophila lamin C is developmentally regulated: analogies with vertebrate A-type lamins. J. Cell Sci. 108, 3189–3198 (1995).
Clines, G. A., Ashley, J. A., Shah, S. & Lovett, M. The structure of the human multiple exostoses 2 gene and characterization of homologs in mouse and Caenorhabditis elegans. Genome Res. 7, 359–367 (1997).
Stickens, D. et al. The EXT2 multiple exostoses gene defines a family of putative tumour suppressor genes. Nature Genet. 14, 25–32 (1996).
Fujiki, Y., Hubbard, A. L., Fowler, S. & Lazarow, P. B. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93, 97–102 (1982).
von Heijne, G. Anew method for predicting signal sequence cleavage sites. Nucleic Acids Res. 14, 4683–90 (1986).
Porter, J. A., Young, K. E. & Beachy, P. A. Cholesterol modification of Hedgehog signaling proteins in animal development. Science 274, 255–259 (1996).
Porter, J. A. et al. Hedgehog patterning activity: role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell 86, 21–34 (1996).
Xu, T. & Rubin, G. M. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117, 1223–1237 (1995).
Nakano, Y. et al. Aprotein with several possible membrane-spanning domains encoded by the Drosophila segment polarity gene patched. Nature 341, 508–513 (1989).
Mullor, J. L., Calleja, M., Capdevila, J. & Guerrero, I. Hedgehog activity, independent of decapentaplegic, participates in wing disc patterning. Development 124, 1227–1237 (1997).
Capdevila, J., Pariente, F., Sampedro, J., Alonso, J. & Guerrero, I. Subcellular localization of the segment polarity protein Patched suggests an interaction with the wingless reception complex in Drosophila embryos. Development 120 (suppl),987–998 (1994).
Taylor, A. M., Nakano, Y., Mohler, J. & Ingham, P. W. Contrasting distributions of patched and Hedgehog proteins in the Drosophila embryo. Mech. Dev. 42, 89–96 (1993).
Dickson, B. J., Dominguez, M., van der Straten, A. & Hafen, E. Control of Drosophila photoreceptor cell fates by phyllopod, a novel nuclear protein acting downstream of the Raf kinase. Cell 80, 453–462 (1995).
Acknowledgements
We thank I. Guerrero, E. Hafen, B. Holmgren, P. Ingham, W. Mothes, G. Struhl and D. Raden for strains or reagents; A. Manoukian for his initial contribution to this work; W. Mothes for help with the microsome assays; B. Seed, C. Tabin and S. Goode for discussions; and S. Goode, B. Mathey-Prevot, S. van den Heuvel, M. Zeidler, E. Bach and M. Petitt for comments on the manuscript. Y.B. thanks A.Morineau for encouragement during this study. Y.B. is supported by the Boheringer Ingelheim Fonds; I.T. is supported by the Damon Runyon Fellowship. This work was supported by the NSF and the HHMI where N.P. is an investigator.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bellaiche, Y., The, I. & Perrimon, N. Tout-velu is a Drosophila homologue of the putative tumour suppressor EXT-1 and is needed for Hh diffusion. Nature 394, 85–88 (1998). https://doi.org/10.1038/27932
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/27932
This article is cited by
-
Hedgehog signaling and its molecular perspective with cholesterol: a comprehensive review
Cellular and Molecular Life Sciences (2022)
-
Association of Sonic Hedgehog with the extracellular matrix requires its zinc-coordination center
BMC Molecular and Cell Biology (2021)
-
Bridging the gap: heparan sulfate and Scube2 assemble Sonic hedgehog release complexes at the surface of producing cells
Scientific Reports (2016)
-
The Hedgehog pathway: role in cell differentiation, polarity and proliferation
Archives of Toxicology (2015)
-
Genetic screening of EXT1 and EXT2 in Cypriot families with hereditary multiple osteochondromas
Journal of Genetics (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.