Abstract
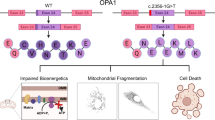
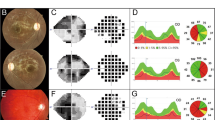
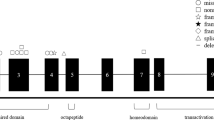
The newly recognized ataxia–ocular apraxia 1 (AOA1; MIM 208920)1,2,3,4 is the most frequent cause of autosomal recessive ataxia in Japan2,4,5,6,7,8,9 and is second only to Friedreich ataxia in Portugal10. It shares several neurological features with ataxia-telangiectasia, including early onset ataxia, oculomotor apraxia and cerebellar atrophy, but does not share its extraneurological features (immune deficiency, chromosomal instability and hypersensitivity to X-rays). AOA1 is also characterized by axonal motor neuropathy3,5,9 and the later decrease of serum albumin levels and elevation of total cholesterol2,4,5,9. We have identified the gene causing AOA1 and the major Portuguese and Japanese mutations. This gene encodes a new, ubiquitously expressed protein that we named aprataxin. This protein is composed of three domains that share distant homology with the amino-terminal domain of polynucleotide kinase 3′- phosphatase (PNKP), with histidine-triad (HIT) proteins and with DNA-binding C2H2 zinc-finger proteins, respectively. PNKP is involved in DNA single-strand break repair (SSBR)11 following exposure to ionizing radiation and reactive oxygen species. Fragile-HIT proteins (FHIT) cleave diadenosine tetraphosphate, which is potentially produced during activation of the SSBR complex12. The results suggest that aprataxin is a nuclear protein with a role in DNA repair reminiscent of the function of the protein defective in ataxia-telangiectasia, but that would cause a phenotype restricted to neurological signs when mutant.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Aicardi, J. et al. Ataxia-ocular motor apraxia: a syndrome mimicking ataxia-telangiectasia. Ann. Neurol. 24, 497–502 (1988).
Uekawa, K., Yuasa, T., Kawasaki, S., Makibuchi, T. & Ideta, T. A hereditary ataxia associated with hypoalbuminemia and hyperlipidemia—a variant form of Friedreich's disease or a new clinical entity? Clin. Neurol. 32, 1067–1074 (1992).
Barbot, C. et al. Recessive ataxia with ocular apraxia: review of 22 Portuguese patients. Arch. Neurol. 58, 201–205 (2001).
Moreira, M.C. et al. Homozygosity mapping of Portuguese and Japanese forms of ataxia-oculomotor apraxia to 9p13, and evidence for genetic heterogeneity. Am. J. Hum. Genet. 68, 501–508 (2001).
Fukuhara, N., Nakajima, T., Sakajiri, K., Matsubara, N. & Fujita, M. Hereditary motor and sensory neuropathy associated with cerebellar atrophy (HMSNCA): a new disease. J. Neurol. Sci. 133, 140–151 (1995).
Hanihara, T., Kubota, H., Amano, N., Iwamoto, H. & Iwabuchi, K. Siblings of early onset cerebellar ataxia with hypoalbuminemia. Rinsho Shinkeigaku (Clin. Neurol.) 35, 83–86 (1995).
Kubota, H. et al. Familial early onset cerebellar ataxia with hypoalbuminemia. No To Shinkei 47, 289–294 (1995).
Sekijima, Y. et al. Hereditary motor and sensory neuropathy associated with cerebellar atrophy (HMSNCA): clinical and neuropathological features of a Japanese family. J. Neurol. Sci. 158, 30–37 (1998).
Tachi, N., Kozuka, N., Ohya, K., Chiba, S. & Sasaki, K. Hereditary cerebellar ataxia with peripheral neuropathy and mental retardation. Eur. Neurol. 43, 82–87 (2000).
Moreira, M.C., Miranda, C., Barbot, C., Coutinho, P. & Sequeiros, J. Recessive ataxias in Portugal. Arq. Med. 13, 39–45 (1999).
Whitehouse, C.J. et al. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell 104, 107–117 (2001).
McLennan, A.G. Dinucleoside polyphosphates—friend or foe? Pharmacol. Ther. 87, 73–89 (2000).
Date, H. et al. Early onset ataxia with ocular–motor apraxia and hypoalbuminemia is caused by mutations in a new HIT-superfamily gene. Nature Genet. 29, 184–188 (2001).
Jilani, A. et al. Molecular cloning of the human gene, PNKP, encoding a polynucleotide kinase 3′-phosphatase and evidence for its role in repair of DNA strand breaks caused by oxidative damage. J. Biol. Chem. 274, 24176–24186 (1999).
Karimi-Busheri, F. et al. Molecular characterization of a human DNA kinase. J. Biol. Chem. 274, 24187–24194 (1999).
Brenner, C., Bieganowski, P., Pace, H.C. & Huebner, K. The histidine triad superfamily of nucleotide-binding proteins. J. Cell. Physiol. 181, 179–187 (1999).
Yoshihara, K. & Tanaka, Y. ADP-ribosylation of diadenosine 5′,5″-P1,P4-tetraphosphate by poly(ADP-ribose) polymerase in vitro. J. Biol. Chem. 256, 6756–6761 (1981).
Caldecott, K.W., Aoufouchi, S., Johnson, P. & Shall, S. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular 'nick-sensor' in vitro. Nucleic Acids. Res. 24, 4387–4394 (1996).
Mackey, Z.B. et al. DNA ligase III is recruited to DNA strand breaks by a zinc finger motif homologous to that of poly(ADP-ribose) polymerase. Identification of two functionally distinct DNA binding regions within DNA ligase III. J. Biol. Chem. 274, 21679–21687 (1999).
Gibson, T.J., Postma, J.P.M., Brown, R.S. & Argos, P. A model for the tertiary structure of the 28 residue DNA-binding motif ('zinc finger') common to many eukaryotic transcriptional regulatory proteins. Protein Engng. 2, 209–218 (1988).
Gatti, R.A. et al. Ataxia-telangiectasia: an interdisciplinary approach to pathogenesis. Medicine 70, 99–117 (1991).
Hawley, R.S. & Friend, S.H. Strange bedfellows in even stranger places: the role of ATM in meiotic cells, lymphocytes, tumors, and its functional links to p53. Genes Dev. 10, 2383–2388 (1996).
Durocher, D. & Jackson, S.P. DNA-PK, ATM and ATR as sensors of DNA damage: variations on a theme? Curr. Opin. Cell Biol. 13, 225–231 (2001).
Kastan, M.B. & Lim, D.S. The many substrates and functions of ATM. Nature Rev. Mol. Cell Biol. 1, 179–186 (2000).
Shiloh, Y. ATM and ATR: networking cellular responses to DNA damage. Curr. Opin. Genet. Dev. 11, 71–77 (2001)
Schultz, J., Copley, R.R., Doerks, T., Ponting, C.P. & Bork, P. SMART: a Web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28, 231–234 (2000).
Acknowledgements
We wish to express our gratitude to all patients and families for their collaboration, as well as to R. Chorão, C. Ferreira, I. Fineza, K. Dias, J.P. Monteiro, K. Sasaki, M. Shizuka-Ikeda, C. Alves, L. Pereira and A. Amorim. We are indebted to J.-L. Mandel for his enthusiastic support and fruitful discussions; to D. Simon, H. Puccio and A. Buj-Bello for their support and their sharing of biological material; and to D. Sommer-Stephan, S. Vicaire, E. Troesch, F. Ruffenach, I. Colas and T. Matamá for excellent technical help. Family characterization and DNA sampling were supported by a grant from the Comissão de Fomento da Investigação em Cuidados de Saúde, Portuguese Ministry of Health (no. 207/99) and grants from Fundação para a Ciência e a Tecnologia (Portuguese Ministry of Science and Technology) and from the Portuguese Ministry of Health (projects PRAXIS/PSAU/P/SAU/84/96, POCTI 34535/99, POCTI 32643/99 and PECS/C/SAU/219/95). Genetic studies were supported by funds from the Institut National de la Santé et de la Recherche Médicale, the Centre National de la Recherche Scientifique, the Hôpitaux Universitaires de Strasbourg, and the Human Frontier Science Program (to M.K. and T.B.). M.C.M. has graduate fellowship PRAXIS XXI/BD/18169/98 from the Fundação para a Ciência e a Tecnologia—Portugal.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Rights and permissions
About this article
Cite this article
Moreira, MC., Barbot, C., Tachi, N. et al. The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nat Genet 29, 189–193 (2001). https://doi.org/10.1038/ng1001-189
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/ng1001-189
This article is cited by
-
A Review of Ocular Movement Abnormalities in Hereditary Cerebellar Ataxias
The Cerebellum (2023)
-
Senescence and impaired DNA damage responses in alpha-synucleinopathy models
Experimental & Molecular Medicine (2022)
-
Signal-on/signal-off bead-based assays for the multiplexed monitoring of base excision repair activities by flow cytometry
Analytical and Bioanalytical Chemistry (2022)
-
HMGB1 signaling phosphorylates Ku70 and impairs DNA damage repair in Alzheimer’s disease pathology
Communications Biology (2021)
-
Conventional MRI findings in hereditary degenerative ataxias: a pictorial review
Neuroradiology (2021)