Key Points
-
The α-catenin family consists of four members — αE-catenin, αN-catenin, αT-catenin and α-catulin — all of which seem to have important functions in development and differentiation.
-
Several interesting insights regarding α-catenin function have been gained from structural studies of the α-catenin M-fragment, dimerization and β-catenin-binding domains, as well as comparisons with vinculin structure.
-
α-catenin seems to be a central component in nucleating the assembly of a multiprotein complex that links E-cadherin–β-catenin complexes to F-actin. This is important for stabilizing adherens junctions, sealing membranes and assembling epithelial sheets. Several proteins link α-catenin to the actin cytoskeleton.
-
The dynamic remodelling of adhesive contacts is required for the maintenance of organized epithelia, so binding of α-catenin to the E-cadherin–β-catenin complex has to be tightly regulated.
-
The disruption of cadherin-based adhesion has a central role in the progression of human epithelial cancers. Reduced levels of E-cadherin and α-catenin proteins have been reported as a characteristic of many different human cancers.
-
α-catenin interacts with proteins, such as Ajuba, which might link adhesion and signalling in the nucleus.
Abstract
α-catenin has often been considered to be a non-regulatory intercellular adhesion protein, in contrast to β-catenin, which has well-documented dual roles in cell–cell adhesion and signal transduction. Recently, however, α-catenin has been found to be important not only in connecting the E-cadherin–β-catenin complex to the actin cytoskeleton, but also in coordinating actin dynamics and inversely correlating cell adhesion with proliferation. As the number of α-catenin-interacting partners increases, intriguing new connections imply even more complex regulatory functions for this protein.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Huber, O. Structure and function of desmosomal proteins and their role in development and disease. Cell Mol. Life Sci. 60, 1872–1890 (2003).
Ishii, K. Greater diversity of desmosomal cadherins. J. Invest. Dermatol. 120, IX–X (2003).
Garrod, D. R., Merritt, A. J. & Nie, Z. Desmosomal cadherins. Curr. Opin. Cell Biol. 14, 537–545 (2002).
Perez-Moreno, M., Jamora, C. & Fuchs, E. Sticky business: orchestrating cellular signals at adherens junctions. Cell 112, 535–548 (2003).
Tepass, U. Adherens junctions: new insight into assembly, modulation and function. Bioessays 24, 690–695 (2002).
Yagi, T. & Takeichi, M. Cadherin superfamily genes: functions, genomic organization, and neurologic diversity. Genes Dev. 14, 1169–1180 (2000).
Giles, R. H., van Es, J. H. & Clevers, H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim. Biophys. Acta 1653, 1–24 (2003).
Lustig, B. & Behrens, J. The Wnt signaling pathway and its role in tumor development. J. Cancer Res. Clin. Oncol. 129, 199–221 (2003).
Kikuchi, A. Tumor formation by genetic mutations in the components of the Wnt signaling pathway. Cancer Sci. 94, 225–229 (2003).
Shimoyama, Y. et al. Cadherin dysfunction in a human cancer cell line: possible involvement of loss of α-catenin expression in reduced cell–cell adhesiveness. Cancer Res. 52, 5770–5774 (1992).
Ewing, C. M. et al. Chromosome-5 suppresses tumorigenicity of PC3 prostate-cancer cells: correlation with reexpression of α-catenin and restoration of E-cadherin function. Cancer Res. 55, 4813–4817 (1995).
Morton, R. A., Ewing, C. M., Nagafuchi, A., Tsukita, S. & Isaacs, W. B. Reduction of E-cadherin levels and deletion of the α-catenin gene in human prostate cancer cells. Cancer Res. 53, 3585–3590 (1993).
Kadowaki, T. et al. E-cadherin and α-catenin expression in human esophageal cancer. Cancer Res. 54, 291–296 (1994).
Bullions, L. C., Notterman, D. A., Chung, L. S. & Levine, A. J. Expression of wild-type α-catenin protein in cells with a mutant α-catenin gene restores both growth regulation and tumor suppressor activities. Mol. Cell. Biol. 17, 4501–4508 (1997).
Moll, R., Mitze, M., Frixen, U. H. & Birchmeier, W. Differential loss of E-cadherin expression in infiltrating ductal and lobular breast carcinomas. Am. J. Pathol. 143, 1731–1742 (1993).
Rimm, D. L., Sinard, J. H. & Morrow, J. S. Reduced α-catenin and E-cadherin expression in breast cancer. Lab. Invest. 72, 506–512 (1995).
Shimazui, T., Giroldi, L. A., Bringuier, P. P., Oosterwijk, E. & Schalken, J. A. Complex cadherin expression in renal cell carcinoma. Cancer Res. 56, 3234–3237 (1996).
Vasioukhin, V., Bauer, C., Degenstein, L., Wise, B. & Fuchs, E. Hyperproliferation and defects in epithelial polarity upon conditional ablation of α-catenin in skin. Cell 104, 605–617 (2001). Conditional ablation of αE-catenin in developing skin causes defects in hair-follicle development and epidermal morphogenesis. Adherens-junction formation, intercellular adhesion and epithelial polarity are all affected. Although differentiation occurs, the epidermis shows hyperproliferation, suprabasal mitoses and multinucleate cells.
Tinkle, C. L., Lechler, T., Pasolli, H. A. & Fuchs, E. Conditional targeting of E-cadherin in skin: insights into hyperproliferative and degenerative responses. Proc. Natl Acad. Sci. USA 13, 552–557 (2004).
Drubin, D. G. & Nelson, W. J. Origins of cell polarity. Cell 84, 335–344 (1996).
Gumbiner, B. M. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell 84, 345–357 (1996).
Orsulic, S. & Peifer, M. An in vivo structure–function study of Armadillo, the β-catenin homologue, reveals both separate and overlapping regions of the protein required for cell adhesion and for Wingless signaling. J. Cell Biol. 134, 1283–1300 (1996).
Simske, J. S. et al. The cell junction protein VAB-9 regulates adhesion and epidermal morphology in C. elegans. Nature Cell Biol. 5, 619–625 (2003).
Nagafuchi, A., Takeichi, M. & Tsukita, S. The 102 kd cadherin-associated protein: similarity to vinculin and posttranscriptional regulation of expression. Cell 65, 849–857 (1991).
Hirano, S., Kimoto, N., Shimoyama, Y., Hirohashi, S. & Takeichi, M. Identification of a neural α-catenin as a key regulator of cadherin function and multicellular organization. Cell 70, 293–301 (1992).
Claverie, J. M. et al. Characterization and chromosomal assignment of a human cDNA encoding a protein related to the murine 102-kDa cadherin-associated protein (α-catenin). Genomics 15, 13–20 (1993).
Janssens, B. et al. αT-catenin: a novel tissue-specific β-catenin-binding protein mediating strong cell–cell adhesion. J. Cell Sci. 114, 3177–3188 (2001).
Zhang, J. S. et al. Identification and chromosomal localization of CTNNAL1, a novel protein homologous to α-catenin. Genomics 54, 149–154 (1998).
Park, B. et al. Association of Lbc Rho guanine nucleotide exchange factor with α-catenin-related protein, α-catulin/CTNNAL1, supports serum response factor activation. J. Biol. Chem. 277, 45361–45370 (2002).
Burridge, K. & Fath, K. Focal contacts: transmembrane links between the extracellular matrix and the cytoskeleton. Bioessays 10, 104–108 (1989).
Ozawa, M., Baribault, H. & Kemler, R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J. 8, 1711–1717 (1989).
Nagafuchi, A. & Takeichi, M. Transmembrane control of cadherin-mediated cell adhesion: a 94 kDa protein functionally associated with a specific region of the cytoplasmic domain of E-cadherin. Cell Regul. 1, 37–44 (1989).
Guenet, J. L., Simon-Chazottes, D., Ringwald, M. & Kemler, R. The genes coding for α and β catenin (Catna1 and Catnb) and plakoglobin (Jup) map to mouse chromosomes 18, 9, and 11, respectively. Mamm. Genome 6, 363–366 (1995).
Herrenknecht, K. et al. The uvomorulin-anchorage protein α-catenin is a vinculin homologue. Proc. Natl Acad. Sci. USA 88, 9156–9160 (1991).
Oda, T. et al. Cloning of the human α-catenin cDNA and its aberrant mRNA in a human cancer cell line. Biochem. Biophys. Res. Commun. 193, 897–904 (1993).
Furukawa, Y. et al. Structure, expression and chromosome assignment of the human catenin (cadherin-associated protein) α1 gene CTNNA1. Cytogenet. Cell Genet. 65, 74–78 (1994).
Adams, C. L. & Nelson, W. J. Cytomechanics of cadherin-mediated cell–cell adhesion. Curr. Opin. Cell Biol. 10, 572–577 (1998).
Yonemura, S., Itoh, M., Nagafuchi, A. & Tsukita, S. Cell-to-cell adherens junction formation and actin filament organization: similarities and differences between non-polarized fibroblasts and polarized epithelial cells. J. Cell Sci. 108, 127–142 (1995).
Vasioukhin, V., Bauer, C., Yin, M. & Fuchs, E. Directed actin polymerization is the driving force for epithelial cell–cell adhesion. Cell 100, 209–219 (2000). During the formation of cadherin-mediated intercellular adhesions, Ca2+ stimulates filopodia, which penetrate and embed into neighbouring cells. E-cadherin complexes cluster at the filopodia tips and generate a two-rowed zipper of embedded puncta. Apposing cell surfaces are clamped by desmosomes, whereas vinculin, zyxin, VASP and MENA are recruited to adhesion zippers by a mechanism that requires α-catenin.
Watabe, M., Nagafuchi, A., Tsukita, S. & Takeichi, M. Induction of polarized cell–cell association and retardation of growth by activation of the E-cadherin catenin adhesion system in a dispersed carcinoma line. J. Cell Biol. 127, 247–256 (1994).
Torres, M. et al. An α-E-catenin gene trap mutation defines its function in preimplantation development. Proc. Natl Acad. Sci. USA 94, 901–906 (1997).
Larue, L., Ohsugi, M., Hirchenhain, J. & Kemler, R. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc. Natl Acad. Sci. USA 91, 8263–8267 (1994).
Uchida, N., Honjo, Y., Johnson, K. R., Wheelock, M. J. & Takeichi, M. The catenin/cadherin adhesion system is localized in synaptic junctions bordering transmitter release zones. J. Cell Biol. 135, 767–779 (1996).
Fannon, A. M. & Colman, D. R. A model for central synaptic junctional complex formation based on the differential adhesive specificities of the cadherins. Neuron 17, 423–434 (1996).
Suzuki, S. C., Inoue, T., Kimura, Y., Tanaka, T. & Takeichi, M. Neuronal circuits are subdivided by differential expression of type-II classic cadherins in postnatal mouse brains. Mol. Cell. Neurosci. 9, 433–447 (1997).
Colman, D. R. Neurites, synapses, and cadherins reconciled. Mol. Cell. Neurosci. 10, 1–6 (1997).
Serafini, T. An old friend in a new home: cadherins at the synapse. Trends Neurosci. 20, 322–323 (1997).
Park, C., Falls, W., Finger, J. H., Longo-Guess, C. M. & Ackerman, S. L. Deletion in Catna2, encoding αN-catenin, causes cerebellar and hippocampal lamination defects and impaired startle modulation. Nature Genet. 31, 279–284 (2002). Shows that mice that are homozygous for the cerebellar deficient folia ( cdf ) mutation are ataxic, and have cerebellar hypoplasia and abnormal lobulation of the cerebellum. The deletion on chromosome 6 includes part of Catna2 , which encodes the αN-catenin protein that links the classic cadherins to the neuronal cytoskeleton.
Janssens, B. et al. Assessment of the CTNNA3 gene encoding human αT-catenin regarding its involvement in dilated cardiomyopathy. Hum. Genet. 112, 227–236 (2003).
Rudiger, M. Vinculin and α-catenin: shared and unique functions in adherens junctions. Bioessays 20, 733–740 (1998).
Izard, T. et al. Vinculin activation by talin through helical bundle conversion. Nature 427, 171–175 (2004).
Johnson, R. P. & Craig, S. W. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature 373, 261–264 (1995).
Menkel, A. R. et al. Characterization of an F-actin-binding domain in the cytoskeletal protein vinculin. J. Cell Biol. 126, 1231–1240 (1994).
McGregor, A., Blanchard, A. D., Rowe, A. J. & Critchley, D. R. Identification of the vinculin-binding site in the cytoskeletal protein α-actinin. Biochem. J. 301, 225–233 (1994).
Kroemker, M., Rudiger, A. H., Jockusch, B. M. & Rudiger, M. Intramolecular interactions in vinculin control α-actinin binding to the vinculin head. FEBS Lett. 355, 259–262 (1994).
Rimm, D. L., Koslov, E. R., Kebriaei, P., Cianci, C. D. & Morrow, J. S. α1(E)-catenin is an actin-binding and-bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc. Natl Acad. Sci. USA 92, 8813–8817 (1995). Shows αE-catenin to be a new actin-binding and -bundling protein, and supports a model in which αE-catenin is responsible for organizing and tethering actin filaments at the zones of E-cadherin-mediated cell–cell contact.
Hazan, R. B., Kang, L., Roe, S., Borgen, P. I. & Rimm, D. L. Vinculin is associated with the E-cadherin adhesion complex. J. Biol. Chem. 272, 32448–32453 (1997).
Huttelmaier, S., Bubeck, P., Rudiger, M. & Jockusch, B. M. Characterization of two F-actin-binding and oligomerization sites in the cell-contact protein vinculin. Eur. J. Biochem. 247, 1136–1142 (1997).
Tempel, M., Goldmann, W. H., Isenberg, G. & Sackmann, E. Interaction of the 47-kDa talin fragment and the 32-kDa vinculin fragment with acidic phospholipids: a computer analysis. Biophys. J. 69, 228–241 (1995).
Weiss, E. E., Kroemker, M., Rudiger, A. H., Jockusch, B. M. & Rudiger, M. Vinculin is part of the cadherin–catenin junctional complex: complex formation between α-catenin and vinculin. J. Cell Biol. 141, 755–764 (1998).
Molony, L. & Burridge, K. Molecular shape and self-association of vinculin and metavinculin. J. Cell. Biochem. 29, 31–36 (1985).
Koslov, E. R., Maupin, P., Pradhan, D., Morrow, J. S. & Rimm, D. L. α-catenin can form asymmetric homodimeric complexes and/or heterodimeric complexes with β-catenin. J. Biol. Chem. 272, 27301–27306 (1997). Reports that α-catenin exists as a homodimer in solution, whereas β-catenin exists as a monomer. When both are present, they form α–β-catenin heterodimers. The site of α-catenin dimerization localizes to the β-catenin-binding site.
Craig, S. W. & Johnson, R. P. Assembly of focal adhesions: progress, paradigms, and portents. Curr. Opin. Cell Biol. 8, 74–85 (1996).
Jockusch, B. M. et al. The molecular architecture of focal adhesions. Annu. Rev. Cell Dev. Biol. 11, 379–416 (1995).
Winkler, J., Lunsdorf, H. & Jockusch, B. M. The ultrastructure of chicken gizzard vinculin as visualized by high-resolution electron microscopy. J. Struct. Biol. 116, 270–277 (1996).
Imamura, Y., Itoh, M., Maeno, Y., Tsukita, S. & Nagafuchi, A. Functional domains of α-catenin required for the strong state of cadherin-based cell adhesion. J. Cell Biol. 144, 1311–1322 (1999).
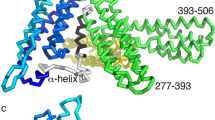
Yang, J., Dokurno, P., Tonks, N. K. & Barford, D. Crystal structure of the M-fragment of α-catenin: implications for modulation of cell adhesion. EMBO J. 20, 3645–3656 (2001). Describes the crystal structure of a region of αE-catenin termed the M-fragment. The region of αE-catenin previously defined as an adhesion M-domain corresponds to the carboxy-terminal four-helix bundle of the M-fragment and these domains exist as dimers in the crystal lattice, which might explain the biological activity of αE-catenin in promoting cell–cell adhesion by inducing lateral dimerization of the associated cadherin molecule.
Yap, A. S., Brieher, W. M., Pruschy, M. & Gumbiner, B. M. Lateral clustering of the adhesive ectodomain: a fundamental determinant of cadherin function. Curr. Biol. 7, 308–315 (1997).
Yap, A. S., Niessen, C. M. & Gumbiner, B. M. The juxtamembrane region of the cadherin cytoplasmic tail supports lateral clustering, adhesive strengthening, and interaction with p120ctn. J. Cell Biol. 141, 779–789 (1998).
Aberle, H. et al. Assembly of the cadherin–catenin complex in vitro with recombinant proteins. J. Cell Sci. 107, 3655–3663 (1994).
Pokutta, S. & Weis, W. I. Structure of the dimerization and β-catenin-binding region of α-catenin. Mol. Cell 5, 533–543 (2000).
Huber, O., Krohn, M. & Kemler, R. A specific domain in α-catenin mediates binding to β-catenin or plakoglobin. J. Cell Sci. 110, 1759–1765 (1997).
Nieset, J. E. et al. Characterization of the interactions of α-catenin with α-actinin and β-catenin/plakoglobin. J. Cell Sci. 110, 1013–1022 (1997).
Obama, H. & Ozawa, M. Identification of the domain of α-catenin involved in its association with β-catenin and plakoglobin (γ-catenin). J. Biol. Chem. 272, 11017–11020 (1997).
Ozawa, M., Ringwald, M. & Kemler, R. Uvomorulin–catenin complex formation is regulated by a specific domain in the cytoplasmic region of the cell adhesion molecule. Proc. Natl Acad. Sci. USA 87, 4246–4250 (1990).
Provost, E. & Rimm, D. L. Controversies at the cytoplasmic face of the cadherin-based adhesion complex. Curr. Opin. Cell Biol. 11, 567–572 (1999).
Watabe-Uchida, M. et al. α-catenin–vinculin interaction functions to organize the apical junctional complex in epithelial cells. J. Cell Biol. 142, 847–857 (1998).
Pokutta, S., Drees, F., Takai, Y., Nelson, W. J. & Weis, W. I. Biochemical and structural definition of the l-afadin- and actin-binding sites of α-catenin. J. Biol. Chem. 277, 18868–18874 (2002).
Ikeda, W. et al. Afadin: a key molecule essential for structural organization of cell–cell junctions of polarized epithelia during embryogenesis. J. Cell Biol. 146, 1117–1132 (1999).
Kobielak, A., Pasolli, H. A. & Fuchs, E. Mammalian formin-1 participates in adherens junctions and polymerization of linear actin cables. Nature Cell Biol. 6, 21–30 (2004).
Wallar, B. J. & Alberts, A. S. The formins: active scaffolds that remodel the cytoskeleton. Trends Cell Biol. 13, 435–446 (2003).
Li, F. & Higgs, H. N. The mouse formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 13, 1335–1340 (2003).
Higashida, C. et al. Actin polymerization-driven molecular movement of mDia1 in living cells. Science 303, 2007–2010 (2004).
Vaezi, A., Bauer, C., Vasioukhin, V. & Fuchs, E. Actin cable dynamics and Rho/Rock orchestrate a polarized cytoskeletal architecture in the early steps of assembling a stratified epithelium. Dev. Cell 3, 367–381 (2002). Reports that during the formation of an epidermal sheet, a polarized network of nascent intercellular junctions and radial actin cables assemble in the apical plane of the monolayer. This polarized cytoskeleton is dependent on α-catenin, Rho and ROCK, and its regulation might be important for wound healing and/or stratification, in which coordinated tissue movements are involved.
Kussel-Andermann, P. et al. Vezatin, a novel transmembrane protein, bridges myosin VIIA to the cadherin–catenins complex. EMBO J. 19, 6020–6029 (2000).
Pradhan, D., Lombardo, C. R., Roe, S., Rimm, D. L. & Morrow, J. S. α-catenin binds directly to spectrin and facilitates spectrin-membrane assembly in vivo. J. Biol. Chem. 276, 4175–4181 (2001).
Ehrlich, J. S., Hansen, M. D. & Nelson, W. J. Spatio-temporal regulation of Rac1 localization and lamellipodia dynamics during epithelial cell–cell adhesion. Dev. Cell 3, 259–270 (2002).
Harden, N. Signaling pathways directing the movement and fusion of epithelial sheets: lessons from dorsal closure in Drosophila. Differentiation 70, 181–203 (2002).
Mullins, R. D. How WASP-family proteins and the Arp2/3 complex convert intracellular signals into cytoskeletal structures. Curr. Opin. Cell Biol. 12, 91–96 (2000).
Adams, C. L., Chen, Y. T., Smith, S. J. & Nelson, W. J. Mechanisms of epithelial cell–cell adhesion and cell compaction revealed by high-resolution tracking of E-cadherin-green fluorescent protein. J. Cell Biol. 142, 1105–1119 (1998).
Evangelista, M., Zigmond, S. & Boone, C. Formins: signaling effectors for assembly and polarization of actin filaments. J. Cell Sci. 116, 2603–2611 (2003).
Hinck, L., Nathke, I. S., Papkoff, J. & Nelson, W. J. Dynamics of cadherin/catenin complex formation: novel protein interactions and pathways of complex assembly. J. Cell Biol. 125, 1327–1340 (1994).
Oda, H., Tsukita, S. & Takeichi, M. Dynamic behavior of the cadherin-based cell–cell adhesion system during Drosophila gastrulation. Dev. Biol. 203, 435–450 (1998).
Daniel, J. M. & Reynolds, A. B. Tyrosine phosphorylation and cadherin/catenin function. Bioessays 19, 883–891 (1997).
Hoschuetzky, H., Aberle, H. & Kemler, R. β-catenin mediates the interaction of the cadherin–catenin complex with epidermal growth factor receptor. J. Cell Biol. 127, 1375–1380 (1994).
Ozawa, M. & Kemler, R. Altered cell adhesion activity by pervanadate due to the dissociation of α-catenin from the E-cadherin–catenin complex. J. Biol. Chem. 273, 6166–6170 (1998).
Braga, V. M. Cell–cell adhesion and signalling. Curr. Opin. Cell Biol. 14, 546–556 (2002).
Fukata, M. et al. Cdc42 and Rac1 regulate the interaction of IQGAP1 with β-catenin. J. Biol. Chem. 274, 26044–26050 (1999).
Kozyraki, R. et al. Expression of cadherins and α-catenin in primary epithelial tumors of the liver. Gastroenterology 110, 1137–1149 (1996).
Ochiai, A. et al. Frequent loss of α-catenin expression in scirrhous carcinomas with scattered cell growth. Jpn. J. Cancer Res. 85, 266–273 (1994).
Shiozaki, H. et al. Immunohistochemical detection of α-catenin expression in human cancers. Am. J. Pathol. 144, 667–674 (1994).
Matsui, S. et al. Immunohistochemical evaluation of α-catenin expression in human gastric cancer. Virchows Arch. 424, 375–381 (1994).
Schipper, J. H. et al. E-cadherin expression in squamous cell carcinomas of head and neck: inverse correlation with tumor dedifferentiation and lymph node metastasis. Cancer Res. 51, 6328–6337 (1991).
Vermeulen, S. J. et al. Transition from the noninvasive to the invasive phenotype and loss of α-catenin in human colon cancer cells. Cancer Res. 55, 4722–4728 (1995).
Gottardi, C. J., Wong, E. & Gumbiner, B. M. E-cadherin suppresses cellular transformation by inhibiting β-catenin signaling in an adhesion-independent manner. J. Cell Biol. 153, 1049–1060 (2001).
Daniel, J. M. & Reynolds, A. B. The catenin p120ctn interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol. Cell. Biol. 19, 3614–3623 (1999).
Marie, H. et al. The LIM protein Ajuba is recruited to cadherin-dependent cell junctions through an association with α-catenin. J. Biol. Chem. 278, 1220–1228 (2003).
Kanungo, J., Pratt, S. J., Marie, H. & Longmore, G. D. Ajuba, a cytosolic LIM protein, shuttles into the nucleus and affects embryonal cell proliferation and fate decisions. Mol. Biol. Cell 11, 3299–3313 (2000).
Zeller, R., Jackson-Grusby, L. & Leder, P. The limb deformity gene is required for apical ectodermal ridge differentiation and anteroposterior limb pattern formation. Genes Dev. 3, 1481–1492 (1989).
Zuniga, A., Haramis, A. P., McMahon, A. P. & Zeller, R. Signal relay by BMP antagonism controls the SHH/FGF4 feedback loop in vertebrate limb buds. Nature 401, 598–602 (1999).
Khokha, M. K., Hsu, D., Brunet, L. J., Dionne, M. S. & Harland, R. M. Gremlin is the BMP antagonist required for maintenance of Shh and Fgf signals during limb patterning. Nature Genet. 34, 303–307 (2003).
Xu, W., Baribault, H., Adamson, E. D. Vinculin knockout results in heart and brain defects during embryonic development. Development 125, 327–337 (1998).
Bakolitsa, C., de Pereda, J. M., Bagshaw, C. R., Critchley, D. R., Liddington, R. C. Crystal structure of the vinculin tail suggests a pathway for activation. Cell 99, 603–613 (1999).
Acknowledgements
We are grateful to D. Barford and D. R. Critchley for providing us with images. E.F. is an investigator at the Howard Hughes Medical Institute, New York, USA. A.K. is a research associate supported by the US National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
Entrez Gene
Swiss-Prot
Glossary
- CLASSIC CADHERINS
-
Cadherins are transmembrane molecules that mediate Ca2+-dependent cell–cell adhesion. Classic cadherins are typified by an extracellular segment that consists of five distinct Ca2+-binding domains and a conserved cytoplasmic domain, which binds β-catenin. The extracellular part interacts homotypically with cadherins on the surface of neighbouring cells to form adherens junctions. The cytoplasmic tail links the actin cytoskeleton to adherens junctions.
- ADHERENS JUNCTION
-
A specialized intercellular junction of the plasma membrane, in which the cadherin molecules of adjacent cells interact in a Ca2+-dependent manner. Actin filaments are linked to this structure through catenins that are located underneath the junction.
- DESMOSOMES
-
Specialized junctional structures that form a tight connection between epithelial cells or cardiac myocytes. They consist of several transmembrane adhesive glycoproteins (desmogleins and desmocollins) and cytoplasmic plaque proteins (desmoplakins) that link to intermediate filaments.
- INTERMEDIATE FILAMENTS
-
Proteins that acquired their name from the diameter of their polymeric structure, which is midway between the diameters of thin actin microfilaments and thick microtubules. Their ability to form very stable filaments enables them to confer mechanical strength on the cytoskeleton.
- ACTOMYOSIN NETWORK
-
A complex of myosin and actin filaments that is responsible for a range of cellular movements in eukaryotic cells. Myosins can translocate vesicles or other cargo on actin filaments.
- TIGHT JUNCTIONS
-
The most apical intercellular junctions, which function as selective (semi-permeable) diffusion barriers between individual cells. They are identified as a belt-like region in which two lipid-apposing membranes lie close together.
- FOCAL ADHESIONS
-
A cell-to-substrate adhesion structure that anchors the ends of actin microfilaments (stress fibres) and mediates strong attachment to substrates.
- GENE TRAP
-
A methodology that is used to characterize new genes and analyse their importance in biological phenomena. The technique involves the use of mouse embryonic stem cells and reporter vectors that are designed to randomly integrate into the genome, tag an insertion site and generate a mutation.
- BLASTOCYST STAGE
-
The stage during embryonic development that is characterized by the formation of two cell types: the embryoblast (the inner cell mass on the inside of the blastocoel) and the trophoblast (the cells on the outside of the blastocoel).
- ACTIVE ZONES
-
The sites along nerve terminals where synaptic vesicles dock and undergo Ca2+-dependent exocytosis during synaptic transmission.
- PURKINJE CELLS
-
Large neurons with extensive dendritic projections that form a layer near to the surface of the cerebellum.
- DILATED CARDIOMYOPATHY
-
Also known as 'congestive cardiomyopathy', this is the most common form of myocardial disease, which causes decreased systolic function and increased ventricular volume.
- AMPHIPATHIC HELICES
-
Helical structures that consist of hydrophobic non-polar residues on one side of the helical cylinder, and hydrophilic and polar residues on the other side.
- YEAST TWO-HYBRID ANALYSIS
-
A technique that is used to study protein–protein interactions in vivo in yeast cells.
- GUANINE NUCLEOTIDE-EXCHANGE FACTOR
-
A protein that facilitates the exchange of guanine diphosphate (GDP) for guanine triphosphate (GTP) in the nucleotide-binding pocket of a GTP-binding protein.
- L CELLS
-
Cells of a mouse fibroblast line that is derived from connective tissue and does not express classic cadherin molecules.
- KERATINOCYTES
-
Differentiated epithelial cells of the skin.
- BARBED END
-
The fast-polymerizing end of an actin filament, which is defined by the arrowhead-shaped decoration of actin filaments with myosin fragments.
- FORMIN FAMILY
-
A family of multidomain scaffold proteins that are involved in actin-dependent morphogenetic events. They are conserved from fungi to humans and are characterized by the presence of two conserved carboxy-terminal regions: the formin homology (FH) domains FH1 and FH2.
- FILOPODIA
-
Thin, transient actin protrusions that extend out from the cell surface and are formed by the elongation of bundled actin filaments in its core.
- LAMELLIPODIA
-
Cellular protrusions that contain extensively branched arrays of actin filaments, which are orientated with their plus (barbed) ends toward the plasma membrane.
- Arp2/3 PROTEIN COMPLEX
-
A complex that consists of two actin-related proteins, Arp2 and Arp3, along with five smaller proteins. When activated, the Arp2/3 complex binds to the side of an existing actin filament and nucleates the assembly of a new actin filament. The resulting branch structure is Y-shaped.
Rights and permissions
About this article
Cite this article
Kobielak, A., Fuchs, E. α-catenin: at the junction of intercellular adhesion and actin dynamics. Nat Rev Mol Cell Biol 5, 614–625 (2004). https://doi.org/10.1038/nrm1433
Issue Date:
DOI: https://doi.org/10.1038/nrm1433
This article is cited by
-
ZO-1 regulates the migration of mesenchymal stem cells in cooperation with α-catenin in response to breast tumor cells
Cell Death Discovery (2024)
-
RNF20 is required for male fertility through regulation of H2B ubiquitination in the Sertoli cells
Cell & Bioscience (2023)
-
Dynamics and functions of E-cadherin complexes in epithelial cell and tissue morphogenesis
Marine Life Science & Technology (2023)
-
Catulin reporter marks a heterogeneous population of invasive breast cancer cells with some demonstrating plasticity and participating in vascular mimicry
Scientific Reports (2022)
-
The intercalated disc: a mechanosensing signalling node in cardiomyopathy
Biophysical Reviews (2020)