-
PDF
- Split View
-
Views
-
Cite
Cite
David J. Finley, Nimmi Arora, Baixin Zhu, Lisa Gallagher, Thomas J. Fahey, Molecular Profiling Distinguishes Papillary Carcinoma from Benign Thyroid Nodules, The Journal of Clinical Endocrinology & Metabolism, Volume 89, Issue 7, 1 July 2004, Pages 3214–3223, https://doi.org/10.1210/jc.2003-031811
Close - Share Icon Share
Abstract
Recently we identified a molecular basis for differentiating benign and malignant follicular thyroid tumors. The purpose of these studies was to determine whether molecular analysis can be used to differentiate papillary thyroid carcinomas from benign thyroid nodules. Gene expression patterns of 14 papillary thyroid carcinomas and 21 benign tumors were analyzed by oligonucleotide array analysis. The carcinomas included seven classical papillary thyroid carcinomas (PTC) and seven follicular variant of PTC (FVPTC), and the benign tumors included 14 follicular adenomas and seven hyperplastic nodules. A hierarchical clustering analysis was performed to examine the groups for potential differences. The combined PTC and FVPTC groups had a distinct gene expression profile compared with the benign lesions. The sensitivity for a diagnosis of carcinoma was 93%, with a 100% specificity (one FVPTC clustered with the benign nodules). Cancer gene profiles contained both known (Met and galectin-3) and previously unidentified genes. Gene profiling is a reliable means of distinguishing PTC, FVPTC, and benign tumors of the thyroid. These gene profiles may provide insight into the pathogenesis of papillary thyroid carcinoma and may ultimately enhance the preoperative diagnosis of thyroid nodules on a molecular basis.
PAPILLARY THYROID CANCER (PTC) is the most common thyroid malignancy, with an estimated incidence of approximately 16,000 cases/yr in the U.S. (1–3). Nevertheless, benign thyroid nodules are far more common, and distinguishing benign and malignant thyroid nodules is a common diagnostic dilemma (4–7). The introduction of fine needle aspiration (FNA) in the 1970s simplified the evaluation of thyroid nodules and decreased the likelihood of surgery for a thyroid nodule by approximately 50% (8). Despite the ability to identify PTC with some reliability on FNA, clinicians frequently have to make a decision regarding the care of patients with thyroid nodules on the basis of equivocal information. For PTC, this diagnostic dilemma is particularly evident in the evaluation of nodules that are potentially follicular variant of papillary thyroid carcinoma (FVPTC) (4, 5). Given such difficulties, these studies were undertaken to analyze PTC, both classical and FVPTC, to determine whether molecular profiling can distinguish these cancers from benign thyroid tumors.
Most large studies have found that between 5 and 29% of thyroid nodules will be deemed a follicular lesion on FNA (6, 9). Many of the nuclear findings associated with PTC, including nuclear grooves, hypochromasia, and pear-shaped or ovoid nuclei, are not readily apparent on cytology due to artifact or sampling of an area that does not include nuclei with those changes (9, 10). Recently, studies evaluating follicular lesions on FNA have found upward of 21% of follicular lesions to be follicular variant of PTC (FVPTC) (5, 7). Baloch et al. (5) noted that 30% of lesions that are FVPTC cannot be diagnosed by FNA. Recent data suggest that FVPTC may be the dominant malignant phenotype of follicular lesions seen in FNA (5, 9, 11). Thus, better techniques are needed to differentiate FVPTC from benign follicular lesions.
Many attempts have been made to identify markers for thyroid carcinoma that can preoperatively distinguish between benign and malignant lesions. Most have not been proven to be specific enough, including PPAR-γ, Met, galectin-3, hTERT, and HBME-1 (12–19). Recently, our laboratory reported the ability to differentiate follicular thyroid carcinoma from follicular adenoma (FA) by means of molecular profiling as well as use this technique to elucidate novel genes that may play an important role in the tumorigenesis of thyroid carcinoma (20). This study reports the ability to differentiate PTC from benign nodules using microarray technology and molecular profiling. This technology may ultimately prove useful in the diagnostic evaluation of thyroid nodules.
Materials and Methods
Tissue procurement
Thyroid tumor tissue was obtained at surgery from patients undergoing thyroidectomy (including hemi, subtotal, and total thyroidectomy). A pathologist dissected the tissue, and a small tumor tissue block from the dominant or suspicious nodule was snap-frozen in liquid nitrogen and stored at −80 C. The size and location of tumor samples were recorded in detail. All tumor samples were obtained with permission of and in accordance with the guidelines of our institutional review board, and informed consent was obtained from all patients. Histological classification was confirmed, the diagnosis was obtained from the final pathology report, and this diagnosis was reviewed and confirmed by an endocrine pathologist.
RNA extraction, purification, labeling, and hybridization
The methods described by Barden et al. (20) were employed for RNA extraction, purification, labeling, and hybridization. In brief, frozen tumor tissue was homogenized by sonication in TRIzol reagent (Invitrogen, Carlsbad, CA), and total RNA was prepared according to the manufacturer’s specifications. A total of 42 samples were analyzed by gene chip array (GeneChip Hu95 array, Affymetrix, Inc., Santa Clara, CA). The carcinoma samples included seven PTC and seven FVPTC. The benign samples consisted of 14 FA and seven hyperplastic nodules. An additional seven unknown samples were processed (blinded to preparer). All samples were processed in the same manner following the Affymetrix protocol. cDNA was synthesized from 8 μg sample RNA using T7 (dT)24 primer (GENSET Corp., La Jolla, CA). Second strand cDNA was then produced and purified. Biotin-labeled cRNA was made and used for hybridization to the Affymetrix oligonucleotide arrays. A sample aliquot was first hybridized to an Affymetrix test chip to confirm that the cRNA quality was adequate. All samples were of good quality. After staining with streptavidin-phycoerythrin, the chips were scanned in an HP ChipScanner (Affymetrix, Inc.) to detect hybridization signals.
Data analysis
The data were analyzed using MicroArray Suite version 5.0 (Affymetrix, Inc.). The intensity of each probe set of the array was captured, and the average intensity was calculated. Quantitative expression levels were calculated using intrachip-positive controls. Normalization of data was performed to account for variability in hybridization among duplicate probe sets and other hybridization artifacts. Transcripts were designated reliably detected (present) or not detected (absent) using the above analysis.
Data analysis was performed to identify genes that were differentially expressed between the papillary carcinoma (PTC and FVPTC) and benign groups (FA and hyperplasia). Data from the 21 benign tumors and 14 carcinomas that comprised the training set were used. First, the data were screened to identify signals counted as present by the Affymetrix software. These results were exported to GeneSpring (Silicon Genetics, Redwood City, CA), then analyzed with a parametric t test and multiple testing correction (Benjamini and Hochberg False Discovery Rate, with the P value set at <0.05), producing a gene list of 1149 genes differentially expressed. This list of 1149 differentially expressed genes was then used for unsupervised hierarchical clustering and statistical analysis. Cluster analysis was used to group the tumors based upon their similarities measured across the expression of 1149 genes.
To determine whether FVPTC could be differentiated from benign thyroid nodules, a second analysis, comparing only FVPTC tumors to benign thyroid nodules, was performed to identify genes differentially expressed between these groups. The data were exported to GeneSpring (Silicon Genetics) and using a nonparametric t test with a P value set at less than 0.01, the data were screened to produce a gene list of 843 differentially expressed genes between FVPTC and benign lesions. Finally, a similar analysis was performed to compare PTC to benign lesions. A total of 483 genes were differentially expressed between PTC and benign lesions. As described above, these gene lists were used for unsupervised hierarchical clustering and statistical analysis.
Evaluation of unknown samples
Once the hierarchical cluster analysis was established using gene expression profiles of differentially expressed genes in 35 tumors, the same analysis was performed on seven thyroid tumors (one PTC, four FVPTC, and two hyperplastic nodules) with investigators blinded to the final diagnosis. Gene profiles of the seven unknown tumors were produced using the 1149 differentially expressed genes for comparison. The unknown sample profiles were then added to the original 35 samples to create a combined group of 42 samples, which then underwent an unsupervised hierarchical clustering analysis.
To confirm the validity of the subset analysis of FVPTC vs. benign tumors, the unknown samples’ individual gene profiles were analyzed (excluding the one classical PTC), using the 843 differentially expressed genes produced by the analysis of FVPTC vs. benign tumors. The gene profiles of unknown samples combined with the test set (FVPTC vs. benign) samples underwent an unsupervised hierarchical cluster analysis, producing a dendrogram for the 34 samples. This same technique was repeated to analyze the PTC vs. benign tumor gene list. Unknown samples’ gene expression profiles were analyzed (excluding the four FVPTC) using the gene profile for 438 genes differentially expressed in the PTC vs. benign analysis. Once again the unknown samples were combined with the PTC vs. benign samples, and an unsupervised hierarchical clustering was performed. A dendrogram of the 31 samples (PTC vs. benign plus unknowns) was produced.
Semiquantitative RT-PCR
To verify the results of the Affymetrix gene chip array, a total of five genes were chosen that had expression levels with more than 2-fold difference, were implicated in the molecular pathogenesis of cancer, and had readily available primers or were made from the known mRNA sequences using Primer3 (21). One microgram of sample RNA was reverse transcribed with oligo(dT) primer in a total volume of 50 μl. A 1-μl aliquot of cDNA was used for PCR, and the product was electrophoresed in a 2.0% agarose gel and visualized with ethidium bromide and UV light. The band intensity for each sample was calculated using EagleSight software (version 3.2, Stratagene, La Jolla, CA), and a ratio of the intensity of the gene of interest to that of the housekeeping gene was calculated for each sample. These normalized intensity levels were then analyzed using a t test. The genes and primers are listed in Table 1.
RT-PCR genes and primers
| . | Sense . | Antisense . | Size (bp) . | Ref. no. . |
|---|---|---|---|---|
| Adrenomedullin | AAGAAGTGGAATAAGTGGGCT | TGGCTTAGAAGACACCAGAGT | 410 | 40 |
| TROP-2 | CCAGTTCCTTGATCTCCACCTTCTT | CTGCTCCACGCTGACCTCCAAGTGT | 753 | 54 |
| MET | GATTTTAGTCATCCCAATGTCC | ATCCAGCATACAGTTTCTTGC | 226 | 55 |
| NRP-2 | CAAACACTGTGGGAACATCG | TTCTCAGGAAACCCAGGAGA | 198 | 43 |
| Trefoil | TTCAAGCCCCTGACTAGGAA | GAGCATGGGACCTTTATTCG | 205 | |
| GAPDH | GAAGGTGAAGGTCGGAGTC | GAAGATGGTGATGGGATTTC | 225 |
| . | Sense . | Antisense . | Size (bp) . | Ref. no. . |
|---|---|---|---|---|
| Adrenomedullin | AAGAAGTGGAATAAGTGGGCT | TGGCTTAGAAGACACCAGAGT | 410 | 40 |
| TROP-2 | CCAGTTCCTTGATCTCCACCTTCTT | CTGCTCCACGCTGACCTCCAAGTGT | 753 | 54 |
| MET | GATTTTAGTCATCCCAATGTCC | ATCCAGCATACAGTTTCTTGC | 226 | 55 |
| NRP-2 | CAAACACTGTGGGAACATCG | TTCTCAGGAAACCCAGGAGA | 198 | 43 |
| Trefoil | TTCAAGCCCCTGACTAGGAA | GAGCATGGGACCTTTATTCG | 205 | |
| GAPDH | GAAGGTGAAGGTCGGAGTC | GAAGATGGTGATGGGATTTC | 225 |
GAPDH, Glyceraldehyde-3-phosphate dehydrogenase
RT-PCR genes and primers
| . | Sense . | Antisense . | Size (bp) . | Ref. no. . |
|---|---|---|---|---|
| Adrenomedullin | AAGAAGTGGAATAAGTGGGCT | TGGCTTAGAAGACACCAGAGT | 410 | 40 |
| TROP-2 | CCAGTTCCTTGATCTCCACCTTCTT | CTGCTCCACGCTGACCTCCAAGTGT | 753 | 54 |
| MET | GATTTTAGTCATCCCAATGTCC | ATCCAGCATACAGTTTCTTGC | 226 | 55 |
| NRP-2 | CAAACACTGTGGGAACATCG | TTCTCAGGAAACCCAGGAGA | 198 | 43 |
| Trefoil | TTCAAGCCCCTGACTAGGAA | GAGCATGGGACCTTTATTCG | 205 | |
| GAPDH | GAAGGTGAAGGTCGGAGTC | GAAGATGGTGATGGGATTTC | 225 |
| . | Sense . | Antisense . | Size (bp) . | Ref. no. . |
|---|---|---|---|---|
| Adrenomedullin | AAGAAGTGGAATAAGTGGGCT | TGGCTTAGAAGACACCAGAGT | 410 | 40 |
| TROP-2 | CCAGTTCCTTGATCTCCACCTTCTT | CTGCTCCACGCTGACCTCCAAGTGT | 753 | 54 |
| MET | GATTTTAGTCATCCCAATGTCC | ATCCAGCATACAGTTTCTTGC | 226 | 55 |
| NRP-2 | CAAACACTGTGGGAACATCG | TTCTCAGGAAACCCAGGAGA | 198 | 43 |
| Trefoil | TTCAAGCCCCTGACTAGGAA | GAGCATGGGACCTTTATTCG | 205 | |
| GAPDH | GAAGGTGAAGGTCGGAGTC | GAAGATGGTGATGGGATTTC | 225 |
GAPDH, Glyceraldehyde-3-phosphate dehydrogenase
Protein purification and Western blotting
Samples were chosen for immunoblot assay based on adequate tumor tissue availability. Frozen tissue was thawed in ice-cold homogenization buffer containing 50 mm Tris-buffered saline (pH 7.4), 2 mm EGTA, 150 mm NaCl, 2 mm MgCl2, 1 mm diethyldithiocarbamate, 1 mm phenylmethylsulfonylfluoride, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 5 μg/ml pepstatin, 3 mm hydrogen peroxide, 50 mm sodium fluoride, 1 mm sodium orthovandate, 10 mm sodium molybdate (Sigma-Aldrich Corp., St. Louis, MO), 10% glycerol (Invitrogen), and 1% Triton-X (Bio-Rad Laboratories, Hercules, CA). Tissues were homogenized using a glass-on-glass tissue homogenizer. Homogenates were centrifuged at 11,750 × g for 10 min at 4 C to remove the particulate material. The protein concentration of the supernatant was measured using the Lowry protein assay kit (Sigma-Aldrich Corp.). Immunoblot analysis for neuropilin-2 (NRP2) was performed using a monoclonal antibody to NRP2 (catalogue no. SC13117, Santa Cruz Biotechnology, Inc., Santa Cruz, CA). One hundred micrograms per lane of protein from tissue were loaded on a 12% SDS-PAGE gel; after transfer, the membrane was blocked in 3% BSA overnight and incubated for 3 h with primary antibody. β-Actin was used as an internal control (Sigma-Aldrich Corp.).
Results
Differentially expressed genes
The level of gene expression in PTC samples (classical and follicular variant) was compared with gene expression in all benign tissue samples (FA and hyperplasia). The resulting gene expression profiles generated a list of differentially expressed genes. A total of 1149 genes were differentially expressed, of which 262 were greater than 2-fold differentially expressed. Seventy-six genes were relatively overexpressed in the carcinoma samples, and 186 genes were relatively overexpressed in the benign tissues (Table 2).
Gene list
| Gene name . | Symbol . | Fold change . | Accession no. . |
|---|---|---|---|
| Overexpressed in carcinomas | |||
| GA733-1 protein precursor | TROP2 | 11.4 | J04152 |
| Similar to LDL receptor-like protein: tSXV motif | MEGF7 | 8.6 | AB011540 |
| Tenascin-C | TNC | 7.9 | X78565 |
| Fibronectin | FN1 | 7.3 | M10905 |
| Dioxin-inducible cytochrome P450 | GLC3A | 7.3 | U03688 |
| α1-Antitrypsin | AAT | 6.9 | X01683 |
| Melanocyte-specific gene 1 | MSG1 | 6.3 | U65092 |
| Intermediate conductance calcium-activated potassium channel | hSK4 | 5.3 | AF022797 |
| Dual specificity protein phosphatase | HVH3 | 5.2 | U15932 |
| NRP-2 | NRP2 | 4.9 | D83018 |
| MAP kinase phosphatase | MKP-2 | 4.7 | U48807 |
| Keratin 19 | KRT19 | 4.7 | Y00503 |
| S plasma protein | PROS | 4.5 | M15036 |
| Galectin-3 | GALBP | 4.4 | AB006780 |
| Glutamine cyclotransferase | GCT | 4.2 | X71125 |
| Tissue inhibitor of metalloproteinases | TIMP | 4.1 | D11139 |
| G Protein-coupled receptor | GABABR2 | 4.0 | AF056085 |
| Human cytokine (GRO-β) | MIP2A | 3.9 | M36820 |
| Precursor alkaline phosphatase | HUSI-I | 3.8 | X04470 |
| TGFα | TGFA | 3.8 | X70340 |
| Signal sequence for secretion by HUVEC | SIG13 | 3.5 | AF015287 |
| Dystrophin | DMD | 3.4 | M18533 |
| KJAA0942 protein | KIAA0942 | 3.4 | AB023159 |
| Lipocortin | LPC1 | 3.3 | X05908 |
| Virally, encoded semaphorin protein receptor | PLXN-C1 | 3.3 | AF030339 |
| Adrenomedullin | AM | 3.3 | D14874 |
| Preprocathepsin H | CPSB | 3.1 | X16832 |
| Breast epithelial antigen 46 | BA46 | 3.0 | U58516 |
| Sky | RSE | 2.9 | D17517 |
| Ryudocan core protein | SYND4 | 2.9 | D79206 |
| Skeletal muscle ryanodine receptor gene | CCO | 2.9 | U48508 |
| Epidermal growth factor receptor kinase substrate | Eps8 | 2.9 | U12535 |
| MET protooncogene protein | HGFR | 2.7 | J02958 |
| HER3 protein precursor | HER3 | 2.7 | M34309 |
| Cathepsin C | CPP1 | 2.6 | X87212 |
| Nuclear factor RIP140 | RIP140 | 2.5 | X84373 |
| TGFβ-stimulated clone 22 | TSC-22 | 2.5 | AJ222700 |
| α1 (VIII) collagen | COL8A1 | 2.5 | X57527 |
| Aryl hydrocarbon receptor | AHR | 2.4 | L19872 |
| SOX-4 protein | SOX-4 | 2.4 | X70683 |
| Aldehyde dehydrogenase | ALDH4 | 2.2 | U10868 |
| Small GTP-binding protein | RAB27 | 2.2 | U57094 |
| N-Acetylglucosamine-6-O-sulfotransferase | GN6ST | 2.2 | AB014679 |
| Single-strand DNA-binding protein | HSPC116 | 2.1 | AL080076 |
| GM3 synthase | SIATGM3S | 2.1 | AB018356 |
| MHC class II HLA-DQ-beta | HLA-DQ-β | 2.1 | M81141 |
| Dihydrouracil dehydrogenase | DPD | 2.1 | U20938 |
| Serine protease hepsin | TMPRSS1 | 2.1 | X07732 |
| Overexpressed in benign tumors | |||
| Collagen α3 type IX | EDM3 | 55.7 | L41162 |
| Intestinal trefoil factor, human | TFF3 | 7.6 | AI985964 |
| Hair keratin | hHb5 | 7.3 | X99140 |
| Creatine kinase B | CKBB | 5.7 | X15334 |
| Thyroid peroxidase | TPX | 5.4 | J02969 |
| Cellular retinoic acid-binding protein-1 | CRABPI | 5.2 | S74445 |
| Thrombospondin-4 | THBS4 | 4.5 | Z19585 |
| IgG Fc-binding protein | FC(γ)BP | 4.1 | D84239 |
| Dystrophin-binding intracellular membrane-associated muscle protein | SNT1 | 4.0 | U40571 |
| Creatine kinase B | CKBB | 3.9 | X15334 |
| Cocaine- and amphetamine-regulated transcript | hCART | 3.9 | U20325 |
| Recombination-activating protein-2 | RAG2 | 3.7 | M94633 |
| GTPase-activating protein | rap1GAP | 3.5 | M64788 |
| Matrilin-2 precursor | MATN2 | 3.5 | U69263 |
| Human metallothionein-9f | MT1 | 3.4 | M10943 |
| SURF-1 | SURF1 | 3.3 | Z35093 |
| Hepatic dihydrodiol dehydrogenase | DDH | 3.3 | U05861 |
| Aldolase A | ALDOA | 3.2 | X05236 |
| Gene name . | Symbol . | Fold change . | Accession no. . |
|---|---|---|---|
| Overexpressed in carcinomas | |||
| GA733-1 protein precursor | TROP2 | 11.4 | J04152 |
| Similar to LDL receptor-like protein: tSXV motif | MEGF7 | 8.6 | AB011540 |
| Tenascin-C | TNC | 7.9 | X78565 |
| Fibronectin | FN1 | 7.3 | M10905 |
| Dioxin-inducible cytochrome P450 | GLC3A | 7.3 | U03688 |
| α1-Antitrypsin | AAT | 6.9 | X01683 |
| Melanocyte-specific gene 1 | MSG1 | 6.3 | U65092 |
| Intermediate conductance calcium-activated potassium channel | hSK4 | 5.3 | AF022797 |
| Dual specificity protein phosphatase | HVH3 | 5.2 | U15932 |
| NRP-2 | NRP2 | 4.9 | D83018 |
| MAP kinase phosphatase | MKP-2 | 4.7 | U48807 |
| Keratin 19 | KRT19 | 4.7 | Y00503 |
| S plasma protein | PROS | 4.5 | M15036 |
| Galectin-3 | GALBP | 4.4 | AB006780 |
| Glutamine cyclotransferase | GCT | 4.2 | X71125 |
| Tissue inhibitor of metalloproteinases | TIMP | 4.1 | D11139 |
| G Protein-coupled receptor | GABABR2 | 4.0 | AF056085 |
| Human cytokine (GRO-β) | MIP2A | 3.9 | M36820 |
| Precursor alkaline phosphatase | HUSI-I | 3.8 | X04470 |
| TGFα | TGFA | 3.8 | X70340 |
| Signal sequence for secretion by HUVEC | SIG13 | 3.5 | AF015287 |
| Dystrophin | DMD | 3.4 | M18533 |
| KJAA0942 protein | KIAA0942 | 3.4 | AB023159 |
| Lipocortin | LPC1 | 3.3 | X05908 |
| Virally, encoded semaphorin protein receptor | PLXN-C1 | 3.3 | AF030339 |
| Adrenomedullin | AM | 3.3 | D14874 |
| Preprocathepsin H | CPSB | 3.1 | X16832 |
| Breast epithelial antigen 46 | BA46 | 3.0 | U58516 |
| Sky | RSE | 2.9 | D17517 |
| Ryudocan core protein | SYND4 | 2.9 | D79206 |
| Skeletal muscle ryanodine receptor gene | CCO | 2.9 | U48508 |
| Epidermal growth factor receptor kinase substrate | Eps8 | 2.9 | U12535 |
| MET protooncogene protein | HGFR | 2.7 | J02958 |
| HER3 protein precursor | HER3 | 2.7 | M34309 |
| Cathepsin C | CPP1 | 2.6 | X87212 |
| Nuclear factor RIP140 | RIP140 | 2.5 | X84373 |
| TGFβ-stimulated clone 22 | TSC-22 | 2.5 | AJ222700 |
| α1 (VIII) collagen | COL8A1 | 2.5 | X57527 |
| Aryl hydrocarbon receptor | AHR | 2.4 | L19872 |
| SOX-4 protein | SOX-4 | 2.4 | X70683 |
| Aldehyde dehydrogenase | ALDH4 | 2.2 | U10868 |
| Small GTP-binding protein | RAB27 | 2.2 | U57094 |
| N-Acetylglucosamine-6-O-sulfotransferase | GN6ST | 2.2 | AB014679 |
| Single-strand DNA-binding protein | HSPC116 | 2.1 | AL080076 |
| GM3 synthase | SIATGM3S | 2.1 | AB018356 |
| MHC class II HLA-DQ-beta | HLA-DQ-β | 2.1 | M81141 |
| Dihydrouracil dehydrogenase | DPD | 2.1 | U20938 |
| Serine protease hepsin | TMPRSS1 | 2.1 | X07732 |
| Overexpressed in benign tumors | |||
| Collagen α3 type IX | EDM3 | 55.7 | L41162 |
| Intestinal trefoil factor, human | TFF3 | 7.6 | AI985964 |
| Hair keratin | hHb5 | 7.3 | X99140 |
| Creatine kinase B | CKBB | 5.7 | X15334 |
| Thyroid peroxidase | TPX | 5.4 | J02969 |
| Cellular retinoic acid-binding protein-1 | CRABPI | 5.2 | S74445 |
| Thrombospondin-4 | THBS4 | 4.5 | Z19585 |
| IgG Fc-binding protein | FC(γ)BP | 4.1 | D84239 |
| Dystrophin-binding intracellular membrane-associated muscle protein | SNT1 | 4.0 | U40571 |
| Creatine kinase B | CKBB | 3.9 | X15334 |
| Cocaine- and amphetamine-regulated transcript | hCART | 3.9 | U20325 |
| Recombination-activating protein-2 | RAG2 | 3.7 | M94633 |
| GTPase-activating protein | rap1GAP | 3.5 | M64788 |
| Matrilin-2 precursor | MATN2 | 3.5 | U69263 |
| Human metallothionein-9f | MT1 | 3.4 | M10943 |
| SURF-1 | SURF1 | 3.3 | Z35093 |
| Hepatic dihydrodiol dehydrogenase | DDH | 3.3 | U05861 |
| Aldolase A | ALDOA | 3.2 | X05236 |
Gene list
| Gene name . | Symbol . | Fold change . | Accession no. . |
|---|---|---|---|
| Overexpressed in carcinomas | |||
| GA733-1 protein precursor | TROP2 | 11.4 | J04152 |
| Similar to LDL receptor-like protein: tSXV motif | MEGF7 | 8.6 | AB011540 |
| Tenascin-C | TNC | 7.9 | X78565 |
| Fibronectin | FN1 | 7.3 | M10905 |
| Dioxin-inducible cytochrome P450 | GLC3A | 7.3 | U03688 |
| α1-Antitrypsin | AAT | 6.9 | X01683 |
| Melanocyte-specific gene 1 | MSG1 | 6.3 | U65092 |
| Intermediate conductance calcium-activated potassium channel | hSK4 | 5.3 | AF022797 |
| Dual specificity protein phosphatase | HVH3 | 5.2 | U15932 |
| NRP-2 | NRP2 | 4.9 | D83018 |
| MAP kinase phosphatase | MKP-2 | 4.7 | U48807 |
| Keratin 19 | KRT19 | 4.7 | Y00503 |
| S plasma protein | PROS | 4.5 | M15036 |
| Galectin-3 | GALBP | 4.4 | AB006780 |
| Glutamine cyclotransferase | GCT | 4.2 | X71125 |
| Tissue inhibitor of metalloproteinases | TIMP | 4.1 | D11139 |
| G Protein-coupled receptor | GABABR2 | 4.0 | AF056085 |
| Human cytokine (GRO-β) | MIP2A | 3.9 | M36820 |
| Precursor alkaline phosphatase | HUSI-I | 3.8 | X04470 |
| TGFα | TGFA | 3.8 | X70340 |
| Signal sequence for secretion by HUVEC | SIG13 | 3.5 | AF015287 |
| Dystrophin | DMD | 3.4 | M18533 |
| KJAA0942 protein | KIAA0942 | 3.4 | AB023159 |
| Lipocortin | LPC1 | 3.3 | X05908 |
| Virally, encoded semaphorin protein receptor | PLXN-C1 | 3.3 | AF030339 |
| Adrenomedullin | AM | 3.3 | D14874 |
| Preprocathepsin H | CPSB | 3.1 | X16832 |
| Breast epithelial antigen 46 | BA46 | 3.0 | U58516 |
| Sky | RSE | 2.9 | D17517 |
| Ryudocan core protein | SYND4 | 2.9 | D79206 |
| Skeletal muscle ryanodine receptor gene | CCO | 2.9 | U48508 |
| Epidermal growth factor receptor kinase substrate | Eps8 | 2.9 | U12535 |
| MET protooncogene protein | HGFR | 2.7 | J02958 |
| HER3 protein precursor | HER3 | 2.7 | M34309 |
| Cathepsin C | CPP1 | 2.6 | X87212 |
| Nuclear factor RIP140 | RIP140 | 2.5 | X84373 |
| TGFβ-stimulated clone 22 | TSC-22 | 2.5 | AJ222700 |
| α1 (VIII) collagen | COL8A1 | 2.5 | X57527 |
| Aryl hydrocarbon receptor | AHR | 2.4 | L19872 |
| SOX-4 protein | SOX-4 | 2.4 | X70683 |
| Aldehyde dehydrogenase | ALDH4 | 2.2 | U10868 |
| Small GTP-binding protein | RAB27 | 2.2 | U57094 |
| N-Acetylglucosamine-6-O-sulfotransferase | GN6ST | 2.2 | AB014679 |
| Single-strand DNA-binding protein | HSPC116 | 2.1 | AL080076 |
| GM3 synthase | SIATGM3S | 2.1 | AB018356 |
| MHC class II HLA-DQ-beta | HLA-DQ-β | 2.1 | M81141 |
| Dihydrouracil dehydrogenase | DPD | 2.1 | U20938 |
| Serine protease hepsin | TMPRSS1 | 2.1 | X07732 |
| Overexpressed in benign tumors | |||
| Collagen α3 type IX | EDM3 | 55.7 | L41162 |
| Intestinal trefoil factor, human | TFF3 | 7.6 | AI985964 |
| Hair keratin | hHb5 | 7.3 | X99140 |
| Creatine kinase B | CKBB | 5.7 | X15334 |
| Thyroid peroxidase | TPX | 5.4 | J02969 |
| Cellular retinoic acid-binding protein-1 | CRABPI | 5.2 | S74445 |
| Thrombospondin-4 | THBS4 | 4.5 | Z19585 |
| IgG Fc-binding protein | FC(γ)BP | 4.1 | D84239 |
| Dystrophin-binding intracellular membrane-associated muscle protein | SNT1 | 4.0 | U40571 |
| Creatine kinase B | CKBB | 3.9 | X15334 |
| Cocaine- and amphetamine-regulated transcript | hCART | 3.9 | U20325 |
| Recombination-activating protein-2 | RAG2 | 3.7 | M94633 |
| GTPase-activating protein | rap1GAP | 3.5 | M64788 |
| Matrilin-2 precursor | MATN2 | 3.5 | U69263 |
| Human metallothionein-9f | MT1 | 3.4 | M10943 |
| SURF-1 | SURF1 | 3.3 | Z35093 |
| Hepatic dihydrodiol dehydrogenase | DDH | 3.3 | U05861 |
| Aldolase A | ALDOA | 3.2 | X05236 |
| Gene name . | Symbol . | Fold change . | Accession no. . |
|---|---|---|---|
| Overexpressed in carcinomas | |||
| GA733-1 protein precursor | TROP2 | 11.4 | J04152 |
| Similar to LDL receptor-like protein: tSXV motif | MEGF7 | 8.6 | AB011540 |
| Tenascin-C | TNC | 7.9 | X78565 |
| Fibronectin | FN1 | 7.3 | M10905 |
| Dioxin-inducible cytochrome P450 | GLC3A | 7.3 | U03688 |
| α1-Antitrypsin | AAT | 6.9 | X01683 |
| Melanocyte-specific gene 1 | MSG1 | 6.3 | U65092 |
| Intermediate conductance calcium-activated potassium channel | hSK4 | 5.3 | AF022797 |
| Dual specificity protein phosphatase | HVH3 | 5.2 | U15932 |
| NRP-2 | NRP2 | 4.9 | D83018 |
| MAP kinase phosphatase | MKP-2 | 4.7 | U48807 |
| Keratin 19 | KRT19 | 4.7 | Y00503 |
| S plasma protein | PROS | 4.5 | M15036 |
| Galectin-3 | GALBP | 4.4 | AB006780 |
| Glutamine cyclotransferase | GCT | 4.2 | X71125 |
| Tissue inhibitor of metalloproteinases | TIMP | 4.1 | D11139 |
| G Protein-coupled receptor | GABABR2 | 4.0 | AF056085 |
| Human cytokine (GRO-β) | MIP2A | 3.9 | M36820 |
| Precursor alkaline phosphatase | HUSI-I | 3.8 | X04470 |
| TGFα | TGFA | 3.8 | X70340 |
| Signal sequence for secretion by HUVEC | SIG13 | 3.5 | AF015287 |
| Dystrophin | DMD | 3.4 | M18533 |
| KJAA0942 protein | KIAA0942 | 3.4 | AB023159 |
| Lipocortin | LPC1 | 3.3 | X05908 |
| Virally, encoded semaphorin protein receptor | PLXN-C1 | 3.3 | AF030339 |
| Adrenomedullin | AM | 3.3 | D14874 |
| Preprocathepsin H | CPSB | 3.1 | X16832 |
| Breast epithelial antigen 46 | BA46 | 3.0 | U58516 |
| Sky | RSE | 2.9 | D17517 |
| Ryudocan core protein | SYND4 | 2.9 | D79206 |
| Skeletal muscle ryanodine receptor gene | CCO | 2.9 | U48508 |
| Epidermal growth factor receptor kinase substrate | Eps8 | 2.9 | U12535 |
| MET protooncogene protein | HGFR | 2.7 | J02958 |
| HER3 protein precursor | HER3 | 2.7 | M34309 |
| Cathepsin C | CPP1 | 2.6 | X87212 |
| Nuclear factor RIP140 | RIP140 | 2.5 | X84373 |
| TGFβ-stimulated clone 22 | TSC-22 | 2.5 | AJ222700 |
| α1 (VIII) collagen | COL8A1 | 2.5 | X57527 |
| Aryl hydrocarbon receptor | AHR | 2.4 | L19872 |
| SOX-4 protein | SOX-4 | 2.4 | X70683 |
| Aldehyde dehydrogenase | ALDH4 | 2.2 | U10868 |
| Small GTP-binding protein | RAB27 | 2.2 | U57094 |
| N-Acetylglucosamine-6-O-sulfotransferase | GN6ST | 2.2 | AB014679 |
| Single-strand DNA-binding protein | HSPC116 | 2.1 | AL080076 |
| GM3 synthase | SIATGM3S | 2.1 | AB018356 |
| MHC class II HLA-DQ-beta | HLA-DQ-β | 2.1 | M81141 |
| Dihydrouracil dehydrogenase | DPD | 2.1 | U20938 |
| Serine protease hepsin | TMPRSS1 | 2.1 | X07732 |
| Overexpressed in benign tumors | |||
| Collagen α3 type IX | EDM3 | 55.7 | L41162 |
| Intestinal trefoil factor, human | TFF3 | 7.6 | AI985964 |
| Hair keratin | hHb5 | 7.3 | X99140 |
| Creatine kinase B | CKBB | 5.7 | X15334 |
| Thyroid peroxidase | TPX | 5.4 | J02969 |
| Cellular retinoic acid-binding protein-1 | CRABPI | 5.2 | S74445 |
| Thrombospondin-4 | THBS4 | 4.5 | Z19585 |
| IgG Fc-binding protein | FC(γ)BP | 4.1 | D84239 |
| Dystrophin-binding intracellular membrane-associated muscle protein | SNT1 | 4.0 | U40571 |
| Creatine kinase B | CKBB | 3.9 | X15334 |
| Cocaine- and amphetamine-regulated transcript | hCART | 3.9 | U20325 |
| Recombination-activating protein-2 | RAG2 | 3.7 | M94633 |
| GTPase-activating protein | rap1GAP | 3.5 | M64788 |
| Matrilin-2 precursor | MATN2 | 3.5 | U69263 |
| Human metallothionein-9f | MT1 | 3.4 | M10943 |
| SURF-1 | SURF1 | 3.3 | Z35093 |
| Hepatic dihydrodiol dehydrogenase | DDH | 3.3 | U05861 |
| Aldolase A | ALDOA | 3.2 | X05236 |
Continued
| Protein C precursor . | PROC . | 3.2 . | X02750 . |
|---|---|---|---|
| Human IgG Fc receptor | FCGRT | 3.1 | U12255 |
| Dipeptidyl aminopeptidase-like protein | DPPX | 3.1 | M96860 |
| Angiopoietin-1 | ANG1 | 3.1 | U83508 |
| Inhibitor of DNA binding 1 | Idt | 3.0 | X77956 |
| Metallothionein-1A | MT1 | 2.9 | K01383 |
| Selenocysteine | TXD12 | 2.9 | AF093774 |
| Heart (R)-3-hydroxybutyrate dehydrogenase | BDH | 2.9 | M93107 |
| Glutathione transferase-ζ1 | GSTZ1 | 2.9 | U86529 |
| Metallothionein IG | MT1 | 2.8 | J03910 |
| KIAA0777 gene product | KIAA0777 | 2.8 | AF049884 |
| Metallothionein III | GIFB | 2.8 | M93311 |
| Very-low-density lipoprotein receptor | VLDLR | 2.7 | D16532 |
| Plasma glutamate carboxypeptidase | PGCP | 2.7 | W28330 |
| Protein kinase X1 | PKX1 | 2.7 | X85545 |
| Endoplasmic reticulum-associated amyloid-β-peptide-binding protein | ERAB | 2.7 | AF035555 |
| KIAA0281 gene | KIAA0281 | 2.7 | D87457 |
| 25-Hydroxyvitamin D3 1-α-hydroxylase | PDDR | 2.7 | AB005038 |
| Extracellular proteinase inhibitor homologue | HE4 | 2.6 | X63187 |
| Ribosomal protein L26 | RPL26 | 2.6 | X69392 |
| Human homolog of a mouse imprinted gene, Peg3 | KIAA0287 | 2.6 | AB006625 |
| K1AA0062 gene | KIAA0062 | 2.6 | D31887 |
| Fibrillin-like (S1-5) | FBNL | 2.6 | U03877 |
| Membrane-type matrix metalloproteinase 2 | MT2-MMP | 2.6 | Z48482 |
| Human ATX1 homolog | HAH1 | 2.6 | U70660 |
| Pituitary tumor-transforming gene | PTTG1 | 2.5 | AA203476 |
| Tetranectin (plasminogen-kringle-4-binding protein) | TN | 2.5 | X64559 |
| Human homolog of mouse Tis11d early response gene | ERF2 | 2.5 | X78992 |
| Human selenium-binding protein | HSBP | 2.5 | U29091 |
| Carbonic anhydrase IV | CAIV | 2.5 | M83670 |
| 17β-Hydroxysteroid dehydrogenase type 3 | EDH17B3 | 2.5 | U05659 |
| Skeletal muscle LIM protein 1 | SLIM1 | 2.5 | AF063002 |
| PLA2 receptor precursor | PLA2R | 2.5 | U17034 |
| KIAA0403 mRNA | KIAA0403 | 2.5 | AB007863 |
| Human metallothionein 1E | MTIE | 2.4 | R92331 |
| Metallothionein 1B | MTI | 2.4 | M13485 |
| Monocyte chemotactic protein-2 | MCP-2 | 2.4 | Y16645 |
| A28-RGS14p mRNA | A28-RGS14 | 2.4 | U70426 |
| Type II transmembrane protein: Golga enzyme | TPST2 | 2.4 | AF049891 |
| Secreted cyclophilin-like protein | CYPB | 2.3 | M63573 |
| Nuclear aconitase | ACO2 | 2.3 | NM_001098 |
| Pax8 | PAX8 | 2.3 | X69699 |
| FK506-binding protein | FKBP12 | 2.3 | M34539 |
| Electron transfer flavoprotein β-subunit | ETFB | 2.3 | X71129 |
| Leukemia inhibitory factor receptor | LIFR | 2.3 | X61615 |
| T Complex-associated testis-expressed 1-like 1 | CW-1 | 2.3 | D50663 |
| Mitochondrial ATP synthase c subunit | ATP5G | 2.3 | X69907 |
| Translation initiation factor e1F-2B β-subunit | EIF2B2 | 2.3 | AF035280 |
| Type 1 inositol 1,4,5-trisphosphate receptor | INSP3R1 | 2.3 | D26070 |
| Pepsinogen | PEP | 2.3 | J00287 |
| Associated with mitotic spindle apparatus | DEEPEST | 2.3 | AF063308 |
| Ribonuclease A | RNS1 | 2.3 | D26129 |
| Synaptic glycoprotein 2 | SC2 | 2.2 | AF038958 |
| Myo-inositol monophosphatase 2 | IMPA2 | 2.2 | AF014398 |
| Histone H1x | H1FX | 2.2 | D64142 |
| P43 | EFTU | 2.2 | S75463 |
| Protein kinase Cθ | PKC | 2.2 | L07032 |
| Human phosphogluconate dehydrogenase | hPGDH | 2.2 | U30255 |
| Prolylcarboxypeptidase | PCP | 2.1 | L13977 |
| Phospholipase C | PLC-L | 2.1 | D42108 |
| Cyclophilin isoform | CYP3 | 2.1 | M80254 |
| Phosphatidylinositol 4,5-bisphosphate 5-phosphatase homolog | PIPP | 2.1 | U45975 |
| Phosphoribosylpyrophosphate synthetase | PRS 1 | 2.1 | D00860 |
| Fibroblast growth factor receptor | K-sam-1 | 2.1 | M87770 |
| Cytochrome e-1 | CYC1 | 2.1 | J04444 |
| Megalin | GP330 | 2.1 | U33837 |
| Lysosomal α-glucosidase | LYAG | 2.0 | X55079 |
| Protein C precursor . | PROC . | 3.2 . | X02750 . |
|---|---|---|---|
| Human IgG Fc receptor | FCGRT | 3.1 | U12255 |
| Dipeptidyl aminopeptidase-like protein | DPPX | 3.1 | M96860 |
| Angiopoietin-1 | ANG1 | 3.1 | U83508 |
| Inhibitor of DNA binding 1 | Idt | 3.0 | X77956 |
| Metallothionein-1A | MT1 | 2.9 | K01383 |
| Selenocysteine | TXD12 | 2.9 | AF093774 |
| Heart (R)-3-hydroxybutyrate dehydrogenase | BDH | 2.9 | M93107 |
| Glutathione transferase-ζ1 | GSTZ1 | 2.9 | U86529 |
| Metallothionein IG | MT1 | 2.8 | J03910 |
| KIAA0777 gene product | KIAA0777 | 2.8 | AF049884 |
| Metallothionein III | GIFB | 2.8 | M93311 |
| Very-low-density lipoprotein receptor | VLDLR | 2.7 | D16532 |
| Plasma glutamate carboxypeptidase | PGCP | 2.7 | W28330 |
| Protein kinase X1 | PKX1 | 2.7 | X85545 |
| Endoplasmic reticulum-associated amyloid-β-peptide-binding protein | ERAB | 2.7 | AF035555 |
| KIAA0281 gene | KIAA0281 | 2.7 | D87457 |
| 25-Hydroxyvitamin D3 1-α-hydroxylase | PDDR | 2.7 | AB005038 |
| Extracellular proteinase inhibitor homologue | HE4 | 2.6 | X63187 |
| Ribosomal protein L26 | RPL26 | 2.6 | X69392 |
| Human homolog of a mouse imprinted gene, Peg3 | KIAA0287 | 2.6 | AB006625 |
| K1AA0062 gene | KIAA0062 | 2.6 | D31887 |
| Fibrillin-like (S1-5) | FBNL | 2.6 | U03877 |
| Membrane-type matrix metalloproteinase 2 | MT2-MMP | 2.6 | Z48482 |
| Human ATX1 homolog | HAH1 | 2.6 | U70660 |
| Pituitary tumor-transforming gene | PTTG1 | 2.5 | AA203476 |
| Tetranectin (plasminogen-kringle-4-binding protein) | TN | 2.5 | X64559 |
| Human homolog of mouse Tis11d early response gene | ERF2 | 2.5 | X78992 |
| Human selenium-binding protein | HSBP | 2.5 | U29091 |
| Carbonic anhydrase IV | CAIV | 2.5 | M83670 |
| 17β-Hydroxysteroid dehydrogenase type 3 | EDH17B3 | 2.5 | U05659 |
| Skeletal muscle LIM protein 1 | SLIM1 | 2.5 | AF063002 |
| PLA2 receptor precursor | PLA2R | 2.5 | U17034 |
| KIAA0403 mRNA | KIAA0403 | 2.5 | AB007863 |
| Human metallothionein 1E | MTIE | 2.4 | R92331 |
| Metallothionein 1B | MTI | 2.4 | M13485 |
| Monocyte chemotactic protein-2 | MCP-2 | 2.4 | Y16645 |
| A28-RGS14p mRNA | A28-RGS14 | 2.4 | U70426 |
| Type II transmembrane protein: Golga enzyme | TPST2 | 2.4 | AF049891 |
| Secreted cyclophilin-like protein | CYPB | 2.3 | M63573 |
| Nuclear aconitase | ACO2 | 2.3 | NM_001098 |
| Pax8 | PAX8 | 2.3 | X69699 |
| FK506-binding protein | FKBP12 | 2.3 | M34539 |
| Electron transfer flavoprotein β-subunit | ETFB | 2.3 | X71129 |
| Leukemia inhibitory factor receptor | LIFR | 2.3 | X61615 |
| T Complex-associated testis-expressed 1-like 1 | CW-1 | 2.3 | D50663 |
| Mitochondrial ATP synthase c subunit | ATP5G | 2.3 | X69907 |
| Translation initiation factor e1F-2B β-subunit | EIF2B2 | 2.3 | AF035280 |
| Type 1 inositol 1,4,5-trisphosphate receptor | INSP3R1 | 2.3 | D26070 |
| Pepsinogen | PEP | 2.3 | J00287 |
| Associated with mitotic spindle apparatus | DEEPEST | 2.3 | AF063308 |
| Ribonuclease A | RNS1 | 2.3 | D26129 |
| Synaptic glycoprotein 2 | SC2 | 2.2 | AF038958 |
| Myo-inositol monophosphatase 2 | IMPA2 | 2.2 | AF014398 |
| Histone H1x | H1FX | 2.2 | D64142 |
| P43 | EFTU | 2.2 | S75463 |
| Protein kinase Cθ | PKC | 2.2 | L07032 |
| Human phosphogluconate dehydrogenase | hPGDH | 2.2 | U30255 |
| Prolylcarboxypeptidase | PCP | 2.1 | L13977 |
| Phospholipase C | PLC-L | 2.1 | D42108 |
| Cyclophilin isoform | CYP3 | 2.1 | M80254 |
| Phosphatidylinositol 4,5-bisphosphate 5-phosphatase homolog | PIPP | 2.1 | U45975 |
| Phosphoribosylpyrophosphate synthetase | PRS 1 | 2.1 | D00860 |
| Fibroblast growth factor receptor | K-sam-1 | 2.1 | M87770 |
| Cytochrome e-1 | CYC1 | 2.1 | J04444 |
| Megalin | GP330 | 2.1 | U33837 |
| Lysosomal α-glucosidase | LYAG | 2.0 | X55079 |
Continued
| Protein C precursor . | PROC . | 3.2 . | X02750 . |
|---|---|---|---|
| Human IgG Fc receptor | FCGRT | 3.1 | U12255 |
| Dipeptidyl aminopeptidase-like protein | DPPX | 3.1 | M96860 |
| Angiopoietin-1 | ANG1 | 3.1 | U83508 |
| Inhibitor of DNA binding 1 | Idt | 3.0 | X77956 |
| Metallothionein-1A | MT1 | 2.9 | K01383 |
| Selenocysteine | TXD12 | 2.9 | AF093774 |
| Heart (R)-3-hydroxybutyrate dehydrogenase | BDH | 2.9 | M93107 |
| Glutathione transferase-ζ1 | GSTZ1 | 2.9 | U86529 |
| Metallothionein IG | MT1 | 2.8 | J03910 |
| KIAA0777 gene product | KIAA0777 | 2.8 | AF049884 |
| Metallothionein III | GIFB | 2.8 | M93311 |
| Very-low-density lipoprotein receptor | VLDLR | 2.7 | D16532 |
| Plasma glutamate carboxypeptidase | PGCP | 2.7 | W28330 |
| Protein kinase X1 | PKX1 | 2.7 | X85545 |
| Endoplasmic reticulum-associated amyloid-β-peptide-binding protein | ERAB | 2.7 | AF035555 |
| KIAA0281 gene | KIAA0281 | 2.7 | D87457 |
| 25-Hydroxyvitamin D3 1-α-hydroxylase | PDDR | 2.7 | AB005038 |
| Extracellular proteinase inhibitor homologue | HE4 | 2.6 | X63187 |
| Ribosomal protein L26 | RPL26 | 2.6 | X69392 |
| Human homolog of a mouse imprinted gene, Peg3 | KIAA0287 | 2.6 | AB006625 |
| K1AA0062 gene | KIAA0062 | 2.6 | D31887 |
| Fibrillin-like (S1-5) | FBNL | 2.6 | U03877 |
| Membrane-type matrix metalloproteinase 2 | MT2-MMP | 2.6 | Z48482 |
| Human ATX1 homolog | HAH1 | 2.6 | U70660 |
| Pituitary tumor-transforming gene | PTTG1 | 2.5 | AA203476 |
| Tetranectin (plasminogen-kringle-4-binding protein) | TN | 2.5 | X64559 |
| Human homolog of mouse Tis11d early response gene | ERF2 | 2.5 | X78992 |
| Human selenium-binding protein | HSBP | 2.5 | U29091 |
| Carbonic anhydrase IV | CAIV | 2.5 | M83670 |
| 17β-Hydroxysteroid dehydrogenase type 3 | EDH17B3 | 2.5 | U05659 |
| Skeletal muscle LIM protein 1 | SLIM1 | 2.5 | AF063002 |
| PLA2 receptor precursor | PLA2R | 2.5 | U17034 |
| KIAA0403 mRNA | KIAA0403 | 2.5 | AB007863 |
| Human metallothionein 1E | MTIE | 2.4 | R92331 |
| Metallothionein 1B | MTI | 2.4 | M13485 |
| Monocyte chemotactic protein-2 | MCP-2 | 2.4 | Y16645 |
| A28-RGS14p mRNA | A28-RGS14 | 2.4 | U70426 |
| Type II transmembrane protein: Golga enzyme | TPST2 | 2.4 | AF049891 |
| Secreted cyclophilin-like protein | CYPB | 2.3 | M63573 |
| Nuclear aconitase | ACO2 | 2.3 | NM_001098 |
| Pax8 | PAX8 | 2.3 | X69699 |
| FK506-binding protein | FKBP12 | 2.3 | M34539 |
| Electron transfer flavoprotein β-subunit | ETFB | 2.3 | X71129 |
| Leukemia inhibitory factor receptor | LIFR | 2.3 | X61615 |
| T Complex-associated testis-expressed 1-like 1 | CW-1 | 2.3 | D50663 |
| Mitochondrial ATP synthase c subunit | ATP5G | 2.3 | X69907 |
| Translation initiation factor e1F-2B β-subunit | EIF2B2 | 2.3 | AF035280 |
| Type 1 inositol 1,4,5-trisphosphate receptor | INSP3R1 | 2.3 | D26070 |
| Pepsinogen | PEP | 2.3 | J00287 |
| Associated with mitotic spindle apparatus | DEEPEST | 2.3 | AF063308 |
| Ribonuclease A | RNS1 | 2.3 | D26129 |
| Synaptic glycoprotein 2 | SC2 | 2.2 | AF038958 |
| Myo-inositol monophosphatase 2 | IMPA2 | 2.2 | AF014398 |
| Histone H1x | H1FX | 2.2 | D64142 |
| P43 | EFTU | 2.2 | S75463 |
| Protein kinase Cθ | PKC | 2.2 | L07032 |
| Human phosphogluconate dehydrogenase | hPGDH | 2.2 | U30255 |
| Prolylcarboxypeptidase | PCP | 2.1 | L13977 |
| Phospholipase C | PLC-L | 2.1 | D42108 |
| Cyclophilin isoform | CYP3 | 2.1 | M80254 |
| Phosphatidylinositol 4,5-bisphosphate 5-phosphatase homolog | PIPP | 2.1 | U45975 |
| Phosphoribosylpyrophosphate synthetase | PRS 1 | 2.1 | D00860 |
| Fibroblast growth factor receptor | K-sam-1 | 2.1 | M87770 |
| Cytochrome e-1 | CYC1 | 2.1 | J04444 |
| Megalin | GP330 | 2.1 | U33837 |
| Lysosomal α-glucosidase | LYAG | 2.0 | X55079 |
| Protein C precursor . | PROC . | 3.2 . | X02750 . |
|---|---|---|---|
| Human IgG Fc receptor | FCGRT | 3.1 | U12255 |
| Dipeptidyl aminopeptidase-like protein | DPPX | 3.1 | M96860 |
| Angiopoietin-1 | ANG1 | 3.1 | U83508 |
| Inhibitor of DNA binding 1 | Idt | 3.0 | X77956 |
| Metallothionein-1A | MT1 | 2.9 | K01383 |
| Selenocysteine | TXD12 | 2.9 | AF093774 |
| Heart (R)-3-hydroxybutyrate dehydrogenase | BDH | 2.9 | M93107 |
| Glutathione transferase-ζ1 | GSTZ1 | 2.9 | U86529 |
| Metallothionein IG | MT1 | 2.8 | J03910 |
| KIAA0777 gene product | KIAA0777 | 2.8 | AF049884 |
| Metallothionein III | GIFB | 2.8 | M93311 |
| Very-low-density lipoprotein receptor | VLDLR | 2.7 | D16532 |
| Plasma glutamate carboxypeptidase | PGCP | 2.7 | W28330 |
| Protein kinase X1 | PKX1 | 2.7 | X85545 |
| Endoplasmic reticulum-associated amyloid-β-peptide-binding protein | ERAB | 2.7 | AF035555 |
| KIAA0281 gene | KIAA0281 | 2.7 | D87457 |
| 25-Hydroxyvitamin D3 1-α-hydroxylase | PDDR | 2.7 | AB005038 |
| Extracellular proteinase inhibitor homologue | HE4 | 2.6 | X63187 |
| Ribosomal protein L26 | RPL26 | 2.6 | X69392 |
| Human homolog of a mouse imprinted gene, Peg3 | KIAA0287 | 2.6 | AB006625 |
| K1AA0062 gene | KIAA0062 | 2.6 | D31887 |
| Fibrillin-like (S1-5) | FBNL | 2.6 | U03877 |
| Membrane-type matrix metalloproteinase 2 | MT2-MMP | 2.6 | Z48482 |
| Human ATX1 homolog | HAH1 | 2.6 | U70660 |
| Pituitary tumor-transforming gene | PTTG1 | 2.5 | AA203476 |
| Tetranectin (plasminogen-kringle-4-binding protein) | TN | 2.5 | X64559 |
| Human homolog of mouse Tis11d early response gene | ERF2 | 2.5 | X78992 |
| Human selenium-binding protein | HSBP | 2.5 | U29091 |
| Carbonic anhydrase IV | CAIV | 2.5 | M83670 |
| 17β-Hydroxysteroid dehydrogenase type 3 | EDH17B3 | 2.5 | U05659 |
| Skeletal muscle LIM protein 1 | SLIM1 | 2.5 | AF063002 |
| PLA2 receptor precursor | PLA2R | 2.5 | U17034 |
| KIAA0403 mRNA | KIAA0403 | 2.5 | AB007863 |
| Human metallothionein 1E | MTIE | 2.4 | R92331 |
| Metallothionein 1B | MTI | 2.4 | M13485 |
| Monocyte chemotactic protein-2 | MCP-2 | 2.4 | Y16645 |
| A28-RGS14p mRNA | A28-RGS14 | 2.4 | U70426 |
| Type II transmembrane protein: Golga enzyme | TPST2 | 2.4 | AF049891 |
| Secreted cyclophilin-like protein | CYPB | 2.3 | M63573 |
| Nuclear aconitase | ACO2 | 2.3 | NM_001098 |
| Pax8 | PAX8 | 2.3 | X69699 |
| FK506-binding protein | FKBP12 | 2.3 | M34539 |
| Electron transfer flavoprotein β-subunit | ETFB | 2.3 | X71129 |
| Leukemia inhibitory factor receptor | LIFR | 2.3 | X61615 |
| T Complex-associated testis-expressed 1-like 1 | CW-1 | 2.3 | D50663 |
| Mitochondrial ATP synthase c subunit | ATP5G | 2.3 | X69907 |
| Translation initiation factor e1F-2B β-subunit | EIF2B2 | 2.3 | AF035280 |
| Type 1 inositol 1,4,5-trisphosphate receptor | INSP3R1 | 2.3 | D26070 |
| Pepsinogen | PEP | 2.3 | J00287 |
| Associated with mitotic spindle apparatus | DEEPEST | 2.3 | AF063308 |
| Ribonuclease A | RNS1 | 2.3 | D26129 |
| Synaptic glycoprotein 2 | SC2 | 2.2 | AF038958 |
| Myo-inositol monophosphatase 2 | IMPA2 | 2.2 | AF014398 |
| Histone H1x | H1FX | 2.2 | D64142 |
| P43 | EFTU | 2.2 | S75463 |
| Protein kinase Cθ | PKC | 2.2 | L07032 |
| Human phosphogluconate dehydrogenase | hPGDH | 2.2 | U30255 |
| Prolylcarboxypeptidase | PCP | 2.1 | L13977 |
| Phospholipase C | PLC-L | 2.1 | D42108 |
| Cyclophilin isoform | CYP3 | 2.1 | M80254 |
| Phosphatidylinositol 4,5-bisphosphate 5-phosphatase homolog | PIPP | 2.1 | U45975 |
| Phosphoribosylpyrophosphate synthetase | PRS 1 | 2.1 | D00860 |
| Fibroblast growth factor receptor | K-sam-1 | 2.1 | M87770 |
| Cytochrome e-1 | CYC1 | 2.1 | J04444 |
| Megalin | GP330 | 2.1 | U33837 |
| Lysosomal α-glucosidase | LYAG | 2.0 | X55079 |
Two-way cluster analysis
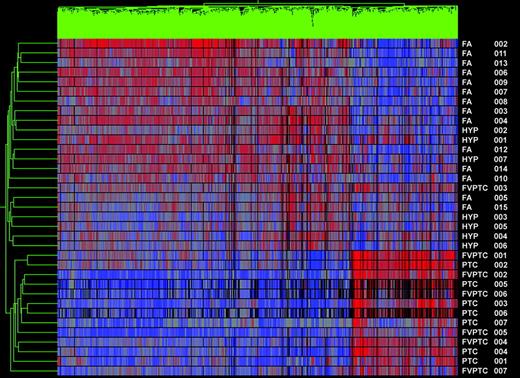
GeneSpring software produces a two-dimensional dendrogram that displays the relatedness of the tumor sample types by both the length and the subdivisions of its clustering. Two distinctly clustered groups are seen in the comparison of PTC (combined PTC and FVPTC) and benign groups (Fig. 1). The top cluster represents only carcinoma samples, whereas the bottom cluster, which represents the benign group, included one FVPTC. Thus, this analysis detected cancer with 93% sensitivity and 100% specificity.
Gene cluster 1. Dendrogram of cluster analysis of 14 PTCs (seven PTC and seven FVPTC) and 21 benign nodules (FAs and hyperplastic nodules) based upon the pattern of expression of 1149 differentially expressed genes. Red indicates relatively high expression; blue indicates low levels of expression. Sample names are listed at the right of the diagram.
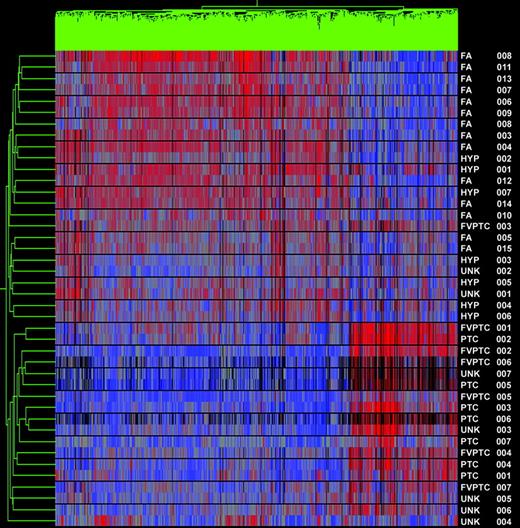
Using the list of differentially expressed genes from the test sets, seven unknown samples were added and assigned to a group based on similarity to other samples by hierarchical cluster analysis. In the combined papillary vs. benign analysis, all seven of the unknown samples were correctly clustered (Fig. 2).
Gene cluster 1 and unknowns. Dendrogram of cluster analysis of 14 PTCs (classic and follicular variant), 21 benign nodules, and seven unknown samples (UNK 1–2 are hyperplastic nodules, UNK 3 is PTC, and UNK 4–7 are FVPTC) based upon the pattern of expression of 1149 differentially expressed genes. Red indicates relatively high expression; blue indicates low levels of expression. All unknowns clustered correctly. Sample names are listed at the right of the diagram.
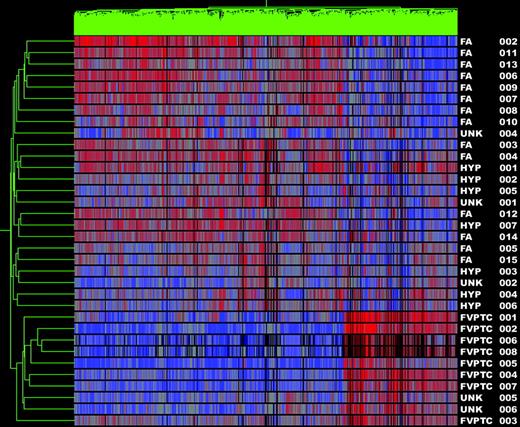
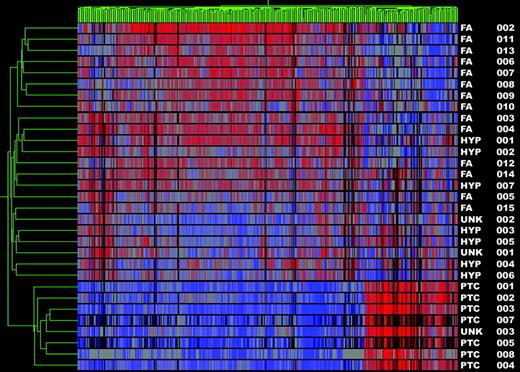
Subset analyses of FVPTC vs. benign nodules and PTC vs. benign nodules was then performed to determine whether the gene profiles could discriminate between the two types of PTC and benign nodules independently. In the analysis of FVPTC vs. benign nodules, 10 of 11 FVPTC clustered together as a distinct cancer group (one FVPTC clustered with the benign nodules). All benign nodules clustered together as a group, yielding a sensitivity of 91% and a specificity of 100% for the diagnosis of cancer (Fig. 3). It is known that FVPTC can be a difficult diagnosis to make even on histopathological review. Thus, a second analysis of the FVPTC vs. benign samples was undertaken using the gene list generated from the classic PTC vs. benign analysis. This second analysis of the FVPTC vs. benign samples revealed results identical to the initial comparison (data not shown). Finally, cluster analysis of PTC vs. benign tumors was 100% sensitive and specific for the diagnosis of cancer (Fig. 4).
Gene cluster 2. Dendrogram of cluster analysis of 11 FVPTC (including three unknowns labeled UNK 4–6) and 23 benign nodules (FAs, hyperplastic nodules, and UNK 1–2) based upon the pattern of expression of 873 differentially expressed genes. Red indicates relatively high expression; blue indicates low levels of expression. Sample names are listed at the right of the diagram.
Gene cluster 3. Dendrogram of cluster analysis of eight PTC (including UNK 3) and 23 benign nodules (FAs, hyperplastic nodules, and UNK 1–2) based upon the pattern of expression of 483 differentially expressed genes. Red indicates relative high expression; blue indicates low levels of expression. Sample names are listed at the right of the diagram.
Corroboration of gene chip analysis
Validation of the gene chip analysis was performed by semiquantitative RT-PCR of five of the most highly differentially expressed genes (Table 2). A total of 26 of the 42 samples were tested, 14 papillary carcinomas (PTC and FVPTC), six FA, and six hyperplasia. The Met oncogene is a gene known to be involved in thyroid tumorigenesis; however, the remaining four genes have not been previously associated with thyroid carcinomas. For each gene tested, the level of expression by RT-PCR correlated with the data obtained by microarray analysis, and all differences seen were statistically significant (P < 0.05; Fig. 5). Additionally, differential expression of the protein NRP2 was confirmed by immunoblot (Fig. 6). The expression of NRP2 was greater in the papillary carcinomas (classic and follicular variant) than in the benign tumors.
Semiquantitative RT-PCR. Verification of highly differentially expressed genes specific for carcinoma or benign tissue. Adrenomedullin (AM), TROP-2, MET, and NRP2 were differentially overexpressed in the carcinomas. Trefoil (intestinal trefoil factor) was relatively overexpressed in benign nodules. Primers specific for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a housekeeping gene, were used as controls. All carcinoma samples (PTC and FVPTC) and 12 of the benign nodules [FA and hyperplastic nodules (HYP)] are displayed. Band intensity levels were normalized to the housekeeping gene and compared using a t test. All differences seen are statistically significant (P < 0.05).
Immunoblot. The differential expression of NRP2 was confirmed by Western blot analysis. One hundred micrograms of protein per lane were loaded. Blot A, The first eight samples (left) are from carcinomas (PTC and FVPTC); the next eight samples (right) are from benign nodules (FA and hyperplastic nodules (HYP)]. Blot B, β-Actin was used as an internal control.
Discussion
PTC constitutes 80% of all differentiated thyroid carcinomas, accounting for almost 16,000 new cases of cancer each year (1–3). Here we have shown that molecular analysis by gene profiling can readily distinguish between PTC, both classical and follicular variant, and benign nodules. These data will provide insight into the molecular pathogenesis of PTCs and may ultimately offer new leads into the treatment of thyroid carcinomas. Additionally, it may provide a molecular basis for the final classification of thyroid tumors that are diagnostic dilemmas for even experienced endocrine pathologists. Finally, it can serve as a springboard for the advancement of diagnosis of thyroid nodules by FNA.
Currently, FNA is sensitive and specific for classic PTC, given the consistency of nuclear findings, including grooving and central clearing (9, 10). However, the ability to define FVPTC by FNA is difficult and limited (5, 9–11). Combining these difficulties with the problems with follicular lesions, FNA is not the optimal preoperative diagnostic tool. Attempts at finding a molecular marker that will readily distinguish benign from malignant thyroid lesions have previously fallen short of the mark. Although many important genetic alterations have been found, including galectin-3, PAX8-PPAR-γ, and MET/HGF receptor, most have not yet proven to be sensitive or specific enough to be translated into clinically useful markers (13, 16–19). Molecular profiling of tumors allows for a quantitative analysis of a panel of genes that are differentially expressed in thyroid nodules, which may lead to the classification of thyroid nodules as either benign or malignant (22, 23). Because of the larger number of genes analyzed, we hypothesized that the profiles generated would overcome the major pitfall of single gene analysis, specifically that in all cases there is overlap of gene expression between benign and malignant tumors. The data presented suggest that our hypothesis is, in fact, true.
Comparison of PTC samples (PTC and FVPTC) with benign lesions by microarray analysis produced a gene list of 262 genes that were significantly differentially expressed. To validate the microarray analysis, we selected five of the 262 genes identified to be differentially expressed between PTC and benign thyroid lesions. Most of these genes have not previously been implicated in thyroid carcinoma pathogenesis and were selected due to their high level of differential expression. Four of these genes were confirmed to be preferentially expressed in PTC, and one was expressed preferentially in the benign tumors. One of the four genes preferentially expressed in papillary carcinoma, the Met gene, has been previously reported to be overexpressed in PTC by both classic molecular techniques and microarray analysis (22, 24). Intestinal trefoil factor, which we found to be overexpressed in the benign group, has previously been reported to be both a tumor suppressor as well as a tumor promoter in other epithelial cell lines (25, 26). The remaining three genes analyzed hold interesting potential as mediators of thyroid carcinogenesis, and these are reviewed below.
TROP2 is a transmembrane receptor protein that has a phosphatidylinositol 4,5-bisphosphate binding sequence on the intracellular portion and is thought to be a receptor-gated calcium channel, integral to calcium homeostasis and possibly cell cycle activation (27–29). Recently, TROP2 has been found to be overexpressed in human epithelial cell tumors, including lung, colon, breast, and ovarian cancers (28–32). Activation of the receptor causes an increase in intracellular calcium levels, and this Ca+ spike is postulated to be a mediator of uncontrolled cell growth (29).
Highly homologous to the cell-cell adhesion molecule TROP1, TROP2 may be involved in cell migration and possibly cell metastasis (29, 33). TROP1 has been used for immunotherapy in colon cancer patients, reducing mortality and relapse rates in resected colon cancer patients, signifying its role in epithelial cell tumors (34, 35). Recently, our laboratory has shown that TROP2 is not highly expressed in follicular thyroid carcinomas (data not shown). Most follicular thyroid carcinomas spread hematogenously, unlike PTC, which mainly metastasizes to lymph nodes (1, 3). Given this distribution of expression between tumors that metastasize differently, TROP2 may be a potential marker for lymphatic metastasis.
Adrenomedullin has been shown to play a role in cancer cell growth and survival (36). A potent angiogenic factor, adrenomedullin, has also been postulated to be a cell mitogen (37). Blocking the activity of adrenomedullin by the use of monoclonal antibodies has shown to suppress tumor cell growth in vitro (38). Recent clinical studies have shown that overexpression of adrenomedullin is correlated with poor prognosis in ovarian cancer (39). Previously our laboratory showed that adrenomedullin was up-regulated in follicular thyroid carcinomas and may be a potential marker for thyroid carcinomas in general (20). Other laboratories have found adrenomedullin abundantly expressed in epithelial tumor cell lines, including lung, breast, ovary, colon, and prostate (40).
Another proangiogenic receptor, NRP2, was found to be overexpressed in PTC. Initially thought only to be a receptor for the semaphorin-3 (SEMA3) family of proteins and expressed during axonal extension, NRP2 has recently been shown to also be a receptor for the vascular endothelial growth factor family of angiogenic factors and has been found to be expressed in NSCLC, osteosarcomas, and pancreatic islet cell and endocrine tumors (41–45). Some postulate that SEMA3F and vascular endothelial growth factor compete for binding to NRP2, and a reduction in SEMA3F leads to increased vascularity, loss of apoptosis, and increased tumor growth (44, 46). Analysis of these three genes (TROP2, adrenomedullin, and NRP2) demonstrates the powerful potential that microarray analysis provides in the identification of new genes that contribute to the pathogenesis of thyroid carcinoma.
The ability to differentiate both classic PTC and FVPTC individually from benign thyroid lesions is clinically significant. Hyperplastic nodules are often difficult to distinguish from FAs on pathological review (47, 48). FVPTC is mistaken for adenomatous lesions, including hyperplasia, on both cytological and histological evaluation, making it difficult to distinguish from benign lesions even on permanent sectioning (47–49). This difficulty has led to the use of multiple immunohistochemical markers, including cytokeratins, to help in differentiating FVPTC from benign lesions (50, 51). Although many markers are positive in FVPTC, they are also positive in benign lesions and ultimately do not significantly increase the sensitivity or specificity in diagnosing these difficult lesions (48, 50, 51). Our ability to differentiate FVPTC from all benign lesions based on the molecular profile with greater than 90% sensitivity and specificity may improve the future accuracy of a diagnosis of cancer.
We have shown in this paper that both classic papillary and the follicular variant of papillary thyroid tumors can be distinguished from benign thyroid nodules based on their molecular profile with excellent sensitivity and specificity. Although classification of tumors based upon their molecular profile is an emerging technology, it has significant clinical potential. Recently, microarray analysis has been performed on FNA biopsies of the breast with hopes of modifying treatment strategies based on FNA molecular analysis (52, 53). Similar potential exists for future preoperative molecular classification of thyroid nodules based on our findings.
This work was supported by a G. Tom Shires Faculty Scholar Award.
Abbreviations:
- FA,
Follicular adenoma;
- FNA,
fine needle aspiration;
- FVPTC,
follicular variant of papillary thyroid carcinoma;
- NRP2,
neuropilin-2;
- PTC,
papillary thyroid carcinoma;
- SEMA3,
semaphorin-3.



![Semiquantitative RT-PCR. Verification of highly differentially expressed genes specific for carcinoma or benign tissue. Adrenomedullin (AM), TROP-2, MET, and NRP2 were differentially overexpressed in the carcinomas. Trefoil (intestinal trefoil factor) was relatively overexpressed in benign nodules. Primers specific for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a housekeeping gene, were used as controls. All carcinoma samples (PTC and FVPTC) and 12 of the benign nodules [FA and hyperplastic nodules (HYP)] are displayed. Band intensity levels were normalized to the housekeeping gene and compared using a t test. All differences seen are statistically significant (P < 0.05).](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/jcem/89/7/10.1210_jc.2003-031811/1/m_zeg0070405850005.jpeg?Expires=1716455693&Signature=f9Hp98suBiWyHG-9MPXIx3NsgvgMtt9AaKmGz0yHY0UxM0HkVoSBhh2e6d50SHgLVqYZtfhsTC~3tnIXz8E9RkAe7qCpRRCZj3vdRRUul-OTQKcq24ZMgK5iJlWi0buez~Wu8z2--iSHZtzvcnFGDADpgHu27UpGaf1TYhnaixdP8DDCO87GRYp~xkmFENdgP3rMHps0sUm1iHSrEXUqD7KhrKK0uwY-aZ-dzZrNpKg2Bzsn323eOGnz~Gd89z8MkaXAH8rgOA~X0FmeYj294IGjptLAci2rzIiuOX3K8xRiFABJqJZDaWKFnlrRN9wresSwIUXt2VkVipGH9cdGVA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Immunoblot. The differential expression of NRP2 was confirmed by Western blot analysis. One hundred micrograms of protein per lane were loaded. Blot A, The first eight samples (left) are from carcinomas (PTC and FVPTC); the next eight samples (right) are from benign nodules (FA and hyperplastic nodules (HYP)]. Blot B, β-Actin was used as an internal control.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/jcem/89/7/10.1210_jc.2003-031811/1/m_zeg0070405850006.jpeg?Expires=1716455693&Signature=xDuvrbYNJNz~QTTosyaNORSlwnm3V1uX4ihVhlSQdsBKzzdOWo6gtslu4aKE9pKI~sq1E57NjtGslQQZ82vbwCWEBQ8fbjOJpYPzbA~Kf4W8pbNoxyxHI0cXaS8lZZRbIIZSWnoxr0ddEmQ7a9VIzSliS1d~wZdSbzkfHDoJcFshHcJB5DP6zKsFx~vaeePaIOAbkL1ISA96pW3hshwTzw8vmTRwEAF~~paKWqLagjnj6h0qP6dLQTmQhZTICBo5hv177bm-0gQKnYfDGp61i~SELYxc-i0XWZRBXg6GWK1ztcrLHJW-ACgdyT4oERaiZ2prdRjdKc-iG0cXFXuDSA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
