Published online Oct 14, 2010. doi: 10.3748/wjg.v16.i38.4858
Revised: June 24, 2010
Accepted: July 1, 2010
Published online: October 14, 2010
AIM: To compare the differences between dideoxy sequencing/KRAS StripAssay/pyrosequencing for detection of KRAS mutation in Chinese colorectal cancer (CRC) patients.
METHODS: Formalin-fixed, paraffin-embedded (FFPE) samples with tumor cells ≥ 50% were collected from 100 Chinese CRC patients at Beijing Cancer Hospital. After the extraction of genome DNA from FFPE samples, fragments contained codons 12 and 13 of KRAS exon 2 were amplified by polymerase chain reaction and analyzed by dideoxy sequencing, the KRAS StripAssay and pyrosequencing. In addition, the sensitivities of the 3 methods were compared on serial dilutions (contents of mutant DNA: 100%, 50%, 20%, 15%, 10%, 5%, 1%, 0%) of A549 cell line DNA (carrying the codon 12 Gly>Ser mutation) into wild-type DNA (human normal intestinal mucosa). The results of dideoxy sequencing, the KRAS StripAssay and pyrosequencing were analyzed by Chromas Software, Collector for KRAS StripAssay and the pyrosequencing PyroMarkTM Q24 system, respectively.
RESULTS: Among 100 patients, KRAS mutations were identified in 34%, 37% and 37% of patients by dideoxy sequencing, the KRAS StripAssay and pyrosequencing, respectively. The sensitivity was highest with the KRAS StripAssay (1%), followed by pyrosequencing (5%), and dideoxy sequencing was lowest (15%). Six different mutation types were found in this study with 3 main mutations Gly12Asp (GGT>GAT), Gly12Val (GGT>GTT) and Gly13Asp (GGC>GAC). Thirty-three patients were identified to have KRAS mutations by the 3 methods, and a total of 8 patients had conflicting results between 3 methods: 4 mutations not detected by dideoxy sequencing and the KRAS StripAssay were identified by pyrosequencing; 3 mutations not detected by dideoxy sequencing and pyrosequencing were identified by the KRAS StripAssay; and 1 mutation not detected by pyrosequencing was confirmed by dideoxy sequencing and the KRAS StripAssay. Among these discordant results, the results identified by dideoxy sequencing were consistent either with the KRAS StripAssay or with pyrosequencing, which indicated that the accuracy of dideoxy sequencing was high.
CONCLUSION: Taking a worldwide view of reports and our results, dideoxy sequencing remains the most popular method because of its low cost and high accuracy.
- Citation: Gao J, Li YY, Sun PN, Shen L. Comparative analysis of dideoxy sequencing, the KRAS StripAssay and pyrosequencing for detection of KRAS mutation. World J Gastroenterol 2010; 16(38): 4858-4864
- URL: https://www.wjgnet.com/1007-9327/full/v16/i38/4858.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i38.4858
Colorectal cancer (CRC) is a common malignant tumor of the gastrointestinal tract, with yearly increasing morbidity and mortality in China. Recently, the development of targeted drugs (for example, cetuximab, panitumumab) has brought advancement in CRC therapy. Cetuximab, which targets the epidermal growth factor receptor (EGFR), shows activity in refractory CRC patients expressing EGFR[1], and in CRC patients with tumors that do not express EGFR immunohistochemically[2]. Thus, there is not an association between EGFR expression and cetuximab efficacy[2,3]. The activating mutations in exon 2 of KRAS play an important role in the progression of CRC, which can induce unlimited proliferation of tumor cells[4,5]. One study reported that KRAS mutations could become an independent prognostic factor in advanced CRC patients treated with cetuximab[6] and there was a significant negative association between KRAS mutations and cetuximab efficacy[7]. Some clinical trials, such as CRYSTAL, OPUS and EVEREST, demonstrated that CRC patients with wild-type KRAS could benefit from the addition of cetuximab to the standard chemotherapy regimen, but patients with mutated KRAS could not[8-10]. Thus, detection of KRAS mutations is strongly recommended before administration of cetuximab.
The mutation rate of KRAS is slightly different among different trials and different areas; for example, the prevalence of KRAS mutation was 35.6% in the CRYSTAL trial[8], but 42.3% in the CO.17 trial[11]; the frequency of KRAS mutation is about 40% in the United States, 34% in the Netherlands, 49% in France and 26.5% in Taiwan, China[12]. The KRAS mutation rate in Chinese CRC patients is about 37% according to our previous study (about 600 patients were studied) using dideoxy sequencing. Most mutations occur in codons 12 and 13 (about 95%), and only a few in codon 61 (about 5%)[13-15]. Up to now, many methods have been used to detect KRAS mutations, including dideoxy sequencing, polymerase chain reaction (PCR)-single-strand conformation polymorphism, PCR-restriction fragment length polymorphism (RFLP), pyrosequencing, denaturing high performance liquid chromatograph, and so on[16]. Along with the advancement of technology, many new kits have been developed, such as the DxS K-RAS Mutation Test Kit and the KRAS StripAssay, which brought new choices for researchers.
As a new method, the KRAS StripAssay has been used in Europe and America, but is not available in China. In order to determine the sensitivity of the KRAS StripAssay, and confirm the feasibility of dideoxy sequencing, we compared the differences between dideoxy sequencing, the KRAS StripAssay and pyrosequencing for mutation detection in codons 12 and 13 of KRAS. Codons 12 and 13 of KRAS were detected by the 3 methods in 100 CRC patients proposed for treatment with cetuximab.
A total of 100 patients in Beijing Cancer Hospital with CRC confirmed by histopathology between October 2008 and August 2009 were investigated for KRAS mutations in our laboratory. Formalin-fixed, paraffin-embedded (FFPE) samples with ≥ 50% tumor cells were collected. An A549 cell line was preserved in our laboratory and normal intestinal mucosa was provided by the tissue bank of our hospital.
Genomic DNA of FFPE sections was extracted using E.Z.N.A.FFPE DNA Kit (Lot. D3399-01, OMEGA, USA) according to the manufacturer’s instructions. Genomic DNAs of A549 and normal intestinal mucosa were extracted using EasyPure Genomic DNA Extraction Kit (Lot. D60916, TransGen Biotech, China) according to the manufacturer’s instructions. All genomic DNAs were stored at -20°C until further research.
The concentrations of DNA from the A549 cell line and normal intestinal mucosa were determined by fluorometry. Serial dilutions were prepared by putting A549 cell DNA into wild-type DNA to produce dilutions with the following contents of mutant DNA: 100%, 50%, 20%, 15%, 10%, 5%, 1%, 0%.
A DNA fragment including exon 2 of the KRAS gene was amplified by PCR using primers (KRAS-F: 5'-GGTACTGGTGGAGTATTTGATAG-3', KRAS-R: 5'-TGGTCCTGCACCAGTAATATG-3') with a product size of 248 bp. Each PCR reaction consisted of 10 × LA PCR buffer II 2 μL, 10 mmol/L dNTPs 2 μL, LA Taq 0.2 μL (DRR200A, TAKARA), genomic DNA 2 μL, 10 μmol/L forward primer 0.5 μL, 10 μmol/L reverse primer 0.5 μL in a final volume of 20 μL. The cycling conditions were 94°C for 5 min, 45 cycles of 94°C for 30 s, 56°C for 30 s and 72°C for 20 s, final extension at 72°C for 10 min, and ended at 4°C. The PCR products were determined by 3% agarose gel electrophoresis and then sequenced using the same forward primer by Invitrogen 3730XL genetic analyzer. The sequencing results were analyzed with Chromas software under the condition of signal/noise > 98%.
The KRAS StripAssay kit (Lot. 5-590) was kindly provided by ViennaLab of Austria. All procedures were conducted according to the manufacturer’s instructions. Briefly, PCR products were amplified in a tube containing 15 μL amplification mix, 5 μL diluted Taq DNA polymerase (1 U), 5 μL DNA template (50 ng genome DNA). The cycling conditions were 94°C for 2 min, 35 cycles of 94°C for 60 s, 70°C for 50 s, 56°C for 50 s and 60°C for 60 s, final extension at 6°C for 3 min. PCR products with fragment lengths 151 bp and 204 bp were determined by 3% agarose gel electrophoresis after PCR amplification. Following hybridization (45°C, shaking waterbath), stringent washing (45°C, shaking waterbath) and color development (room temperature), the results were interpreted using the enclosed Collector sheet.
A DNA fragment including codons 12 and 13 of the KRAS gene was amplified by PCR using primers (forward: 5'-biotin-TGACTGAATATAAACTTGTGGTAGTTG-3', reverse: 5'-TCGTCCACAAAATGATTCTGAA-3') with a product size of 91 bp. Each PCR reaction consisted of 10 × PCR buffer 5 μL, 10 mmol/L dNTPs 4 μL, Hotstart Taq 0.4 μL [Gene Tech (Shanghai) Company Limited], genomic DNA 4 μL, 10 mmol/L forward primer 0.5 μL, 10 mmol/L reverse primer 0.5 μL in a final volume of 50 μL. The cycling conditions were 95°C for 3 min, 45 cycles of 95°C for 10 s, 56°C for 20 s and 72°C for 30 s, final extension at 72°C for 5 min. The PCR products were determined by 3% agarose gel electrophoresis and ssDNA was prepared as described[16]. Mutation detection of KRAS codons 12 and 13 by the Pyrosequencing PyroMark™ Q24 system was done following the manufacturer’s instructions (see http://www.pyrosequencing.com/ for more information).
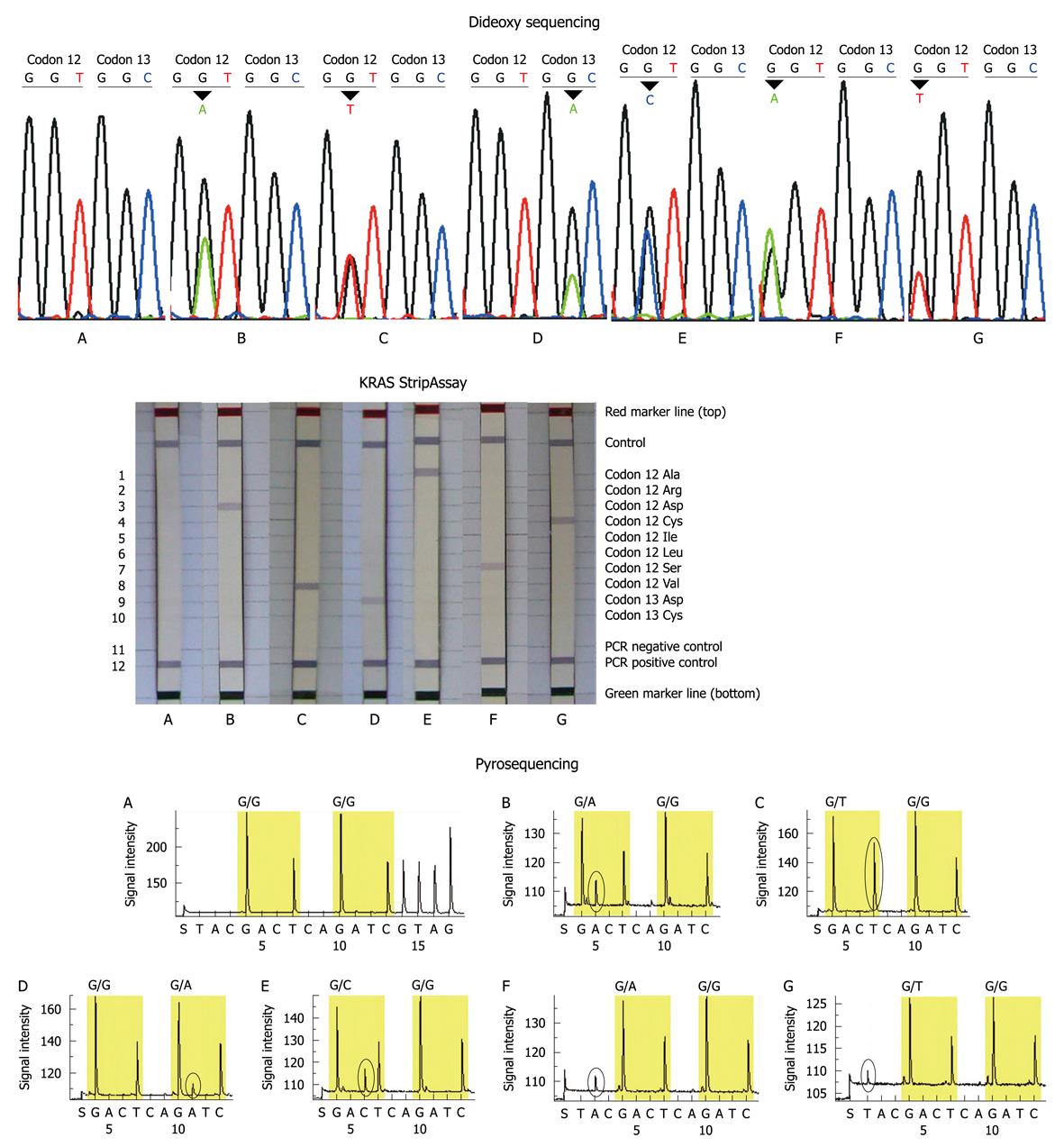
The study included 54 males and 46 females with a median age of 59 years (range 22-82 years). The primary locations of tumors were the colon (n = 58) and rectum (n = 42). The mutation rate in females (about 43%) was slightly higher than that in males (about 30%), and the mutation rate in colon and rectal cancers was similar. All patients had a single mutation site. A total of 6 mutation types were detected in this study: GGT>GAT, GGT>GTT, GTT>GCT, GTT>TGT, GTT>AGT, GGC>GAC (wild-type codon 12: GGT; wild-type codon 13: GGC) (Figure 1). Three main mutations Gly12Asp (GGT>GAT), Gly12Val (GGT>GTT) and Gly13Asp (GGC>GAC) accounted for about 80.0% (28/34) of all mutations.
Serial dilutions with different contents of mutant DNA were detected by the 3 methods. All 3 methods could correctly identify the Gly12Ser mutation in dilutions containing 15% or more mutant DNA. Dideoxy sequencing failed to detect the mutation in dilutions containing 10% or less mutant DNA. Pyrosequencing failed to detect the mutation in dilutions containing 1% mutant DNA, and only the KRAS StripAssay could unambiguously identify 1% mutant DNA in the dilutions. Thus the sensitivity was highest in the KRAS StripAssay (1%), followed by pyrosequencing (5%), while dideoxy sequencing was lowest (15%) (Table 1).
| Methods | Mutant DNA/total DNA (%) | |||||||
| 100 | 50 | 20 | 15 | 10 | 5 | 1 | 0 | |
| Dideoxy sequencing | Yes | Yes | Yes | Yes | No | No | No | No |
| Pyrosequencing | Yes | Yes | Yes | Yes | Yes | Yes | No | No |
| KRAS StripAssay | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No |
The KRAS mutation was detected in 34/100 (34%) of patients by dideoxy sequencing with 15/54 (27.8%) of males and 19/46 (41.3%) of females, 37/100 (37%) of patients by KRAS StripAssay with 16/54 (29.6%) of males and 21/46 (45.7%) of females, and in 37/100 (37%) of patients by pyrosequencing with 18/54 (33.3%) of males and 19/46 (41.3%) of females. Three main mutations Gly12Asp, Gly12Val and Gly13Asp accounted for 82.4% (28/34), 78.4% (29/37) and 83.8% (31/37) of all mutations by dideoxy sequencing, KRAS StripAssay and pyrosequencing, respectively. The overall results of the 3 methods were similar, with a few discrepancies.
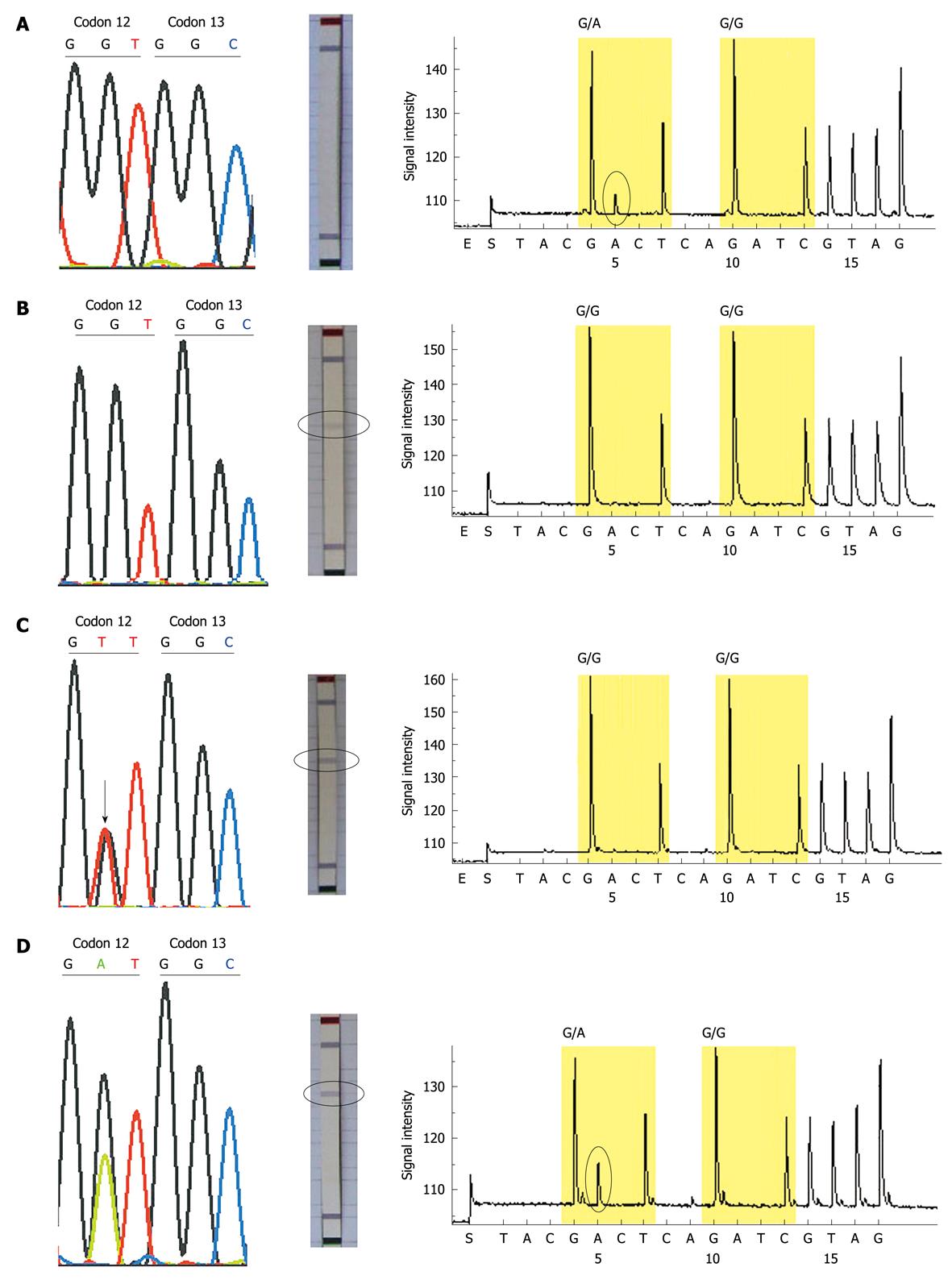
Thirty-three of the 100 patients were identified to have KRAS mutations by all 3 methods, and 8 patients (sample No. 5, 11, 14, 29, 44, 46, 48, 71) showed conflicting results between the 3 methods: 4 mutations (sample No. 5, 44, 46, 48) which were not detected by dideoxy sequencing and the KRAS StripAssay, were identified by pyrosequencing; 3 mutations (sample No. 11, 14, 71) which were not detected by dideoxy sequencing and pyrosequencing were identified by the KRAS StripAssay; one mutation (sample No. 29) not detected by pyrosequencing was idenified by dideoxy sequencing and the KRAS StripAssay (Figure 2). In addition, among these discordant results, the mutations identified by dideoxy sequencing were consistent either with the KRAS StripAssay or with pyrosequencing (Table 2). This indicated that although the sensitivity of dideoxy sequencing was low, its accuracy was high.
| Sample No. | Mutation type | ||
| Dideoxy sequencing | KRAS StripAssay | Pyrosequencing | |
| 5 | Wild-type | Wild-type | GGT>GAT |
| 11 | Wild-type | GGT>TGT | Wild-type |
| 14 | Wild-type | GGT>GTT | Wild-type |
| 29 | GGT>GTT | GGT>GTT | Wild-type |
| 44 | Wild-type | Wild-type | GGT>GAT |
| 46 | Wild-type | Wild-type | GGT>GAT |
| 48 | Wild-type | Wild-type | GGT>GAT |
| 71 | Wild-type | GGT>GCT | Wild-type |
Along with national development and improvements in standard of living, morbidity and mortality of CRC has increased rapidly in China. The outcomes of the same treatment regimen in CRC patients were frequently found to differ, and thus it was important to develop individualized treatments. Because the effect of cetuximab was tightly associated with KRAS mutation status, the US Food and Drug Administration recommended that patients who were proposed for cetuximab treatment should undergo KRAS mutation analysis. As a result, besides the conventional methods, more and more techniques have been developed to detect KRAS mutation.
Recent reports have highlighted the advantages of the new methods, and we first compared dideoxy sequencing, the KRAS StripAssay and pyrosequencing for mutation detection in codons 12 and 13 of KRAS in Chinese CRC patients. The mutation rate of KRAS in our study was about 37%, and the mutation rate in females (about 43%) was higher than in males (about 30%) which was different from a report that KRAS mutation in males was higher than in females in Brazil[16].
The results from the 3 methods were similar, but a total of 8 patients had conflicting results between the 3 methods. We repeated these discrepant samples at least twice, the results being consistent. Because the results by dideoxy sequencing were consistent either with the KRAS StripAssay or with pyrosequencing, the results by dideoxy sequencing were likely to be more accurate. To support our hypothesis, we retrospectively analyzed the patients who were treated with cetuximab. Case 44 with KRAS mutation identified by pyrosequencing was treated with cetuximab and achieved a partial response after 6 weeks’ treatment. Because patients with KRAS mutations could not benefit from cetuximab, the result of case 44 by pyrosequencing may be a false positive.
Our results showed that the sensitivities of the KRAS StripAssay and pyrosequencing were higher than that of dideoxy sequencing, but according to our large-scale sampling by dideoxy sequencing, the KRAS mutation rate was stable at 37-39% which was consistent with other reports. From the result of case 44, a false positive could occur in sensitive methods. We can analyze the 3 methods from the aspect of medical economics. At present, the cost is about 100-150 RMB/test for dideoxy sequencing, 1000 RMB/test for the KRAS StripAssay and 200-300 RMB/test for pyrosequencing. Payments in China are limited to the cheapest methods. Up to now, dideoxy sequencing and pyrosequencing have already been widely used to detect KRAS mutations[17,18], but the KRAS StripAssay has not been widely used all over the world.
Studies reported that the KRAS mutation could be used as a prognostic marker in non-small cell lung cancer and as an independent prognostic factor for CRC patients treated with cetuximab[6,19]. Whether there is a relationship between KRAS mutations and prognosis in Chinese CRC patients needs to be studied further.
In conclusion, we compared the differences between 3 methods in the detection of KRAS mutations, and used the KRAS StripAssay to detect KRAS mutations. Although new methods have been developed for detection of KRAS mutation, traditional methods are still in an invincible position and are used widely. In our following large-scale study, dideoxy sequencing will be chosen preferentially because of its ease of use.
At present, more and more colorectal cancer (CRC) patients are treated with cetuximab [a monoclonal antibody of epidermal growth factor receptor (EGFR)]. It had been confirmed that KRAS mutated patients could not benefit from cetuximab, so the US Food and Drug Administration recommended that patients proposed for cetuximab treatment should undergo KRAS mutation analysis.
Besides the traditional methods (e.g. dideoxy sequencing), more and more methods (various kinds of kits) have been developed to detect KRAS mutation. The authors compared methods and demonstrated differences between dideoxy sequencing, the KRAS StripAssay and pyrosequencing, with indications that although dideoxy sequencing is a traditional method, it is still in an invincible position because of its low cost and high accuracy.
Recently, many reports have highlighted the advantages of new methods, and disregarded the traditional methods in the detection of KRAS mutation. This was the first study to compare the differences between dideoxy sequencing, the KRAS StripAssay and pyrosequencing in KRAS detection. The study indicated that although the sensitivity of dideoxy sequencing was lower than the other 2 methods, it was still widely used by many researchers because of its superior accuracy.
The study could help researchers to choose a suitable method for detection of KRAS mutation according to the sample size, equipment platform and economic status, etc. In their opinion, dideoxy sequencing could be easily carried out in any laboratory.
In normal physiological conditions, KRAS is regulated by its upstream protein EGFR and plays an important role in the development and progression of tumors. Cetuximab could inhibit the tumors through blocking EGFR and the downstream signal pathway. If KRAS was mutated, KRAS protein was activated without the regulation of EGFR, so cetuximab could not have an effect.
The paper deals with the comparison of 3 DNA sequencing methods in order to detect KRAS mutations. The authors postulate that dideoxy sequencing and pyrosequencing techniques have already been widely used to detect KRAS mutations but the KRAS StripAssay has not been widely used all over the world. The paper is well written and well documented.
Peer reviewers: Dr. T Choli-Papadopoulou, Associate Professor, Aristotle University of Thessaloniki, School of Chemistry, Department of Biochmeistry, Thessaloniki, 55124, Greece; Raquel Almeida, PhD, Instituto de Patologia e Imunologia Molecular da Universidade do Porto, Rua Dr Roberto Frias s/n, Porto 4200, Portugal; Dr. Thomas Wex, PD, Clinic of Gastroenterology, Hepatology and Infectious Diseases, Otto-von-Guericke University Magdeburg, Leipziger Str. 44, Magdeburg, 39120, Germany
S- Editor Tian L L- Editor Cant MR E- Editor Zheng XM
| 1. | Saltz LB, Meropol NJ, Loehrer PJ Sr, Needle MN, Kopit J, Mayer RJ. Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J Clin Oncol. 2004;22:1201-1208. [Cited in This Article: ] |
| 2. | Chung KY, Shia J, Kemeny NE, Shah M, Schwartz GK, Tse A, Hamilton A, Pan D, Schrag D, Schwartz L. Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J Clin Oncol. 2005;23:1803-1810. [Cited in This Article: ] |
| 3. | Hebbar M, Wacrenier A, Desauw C, Romano O, Cattan S, Triboulet JP, Pruvot FR. Lack of usefulness of epidermal growth factor receptor expression determination for cetuximab therapy in patients with colorectal cancer. Anticancer Drugs. 2006;17:855-857. [Cited in This Article: ] |
| 4. | Forrester K, Almoguera C, Han K, Grizzle WE, Perucho M. Detection of high incidence of K-ras oncogenes during human colon tumorigenesis. Nature. 1987;327:298-303. [Cited in This Article: ] |
| 5. | Pretlow TP, Brasitus TA, Fulton NC, Cheyer C, Kaplan EL. K-ras mutations in putative preneoplastic lesions in human colon. J Natl Cancer Inst. 1993;85:2004-2007. [Cited in This Article: ] |
| 6. | Lièvre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, Ychou M, Bouché O, Landi B, Louvet C. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26:374-379. [Cited in This Article: ] |
| 7. | Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757-1765. [Cited in This Article: ] |
| 8. | Van Cutsem E, Lang I, D'haens G, Moiseyenko V, Zaluski J, Folprecht G, Tejpar S, Kisker O, Stroh C, Rougier P. KRAS status and efficacy in the first-line treatment of patients with metastatic colorectal cancer (mCRC) treated with FOLFIRI with or without cetuximab: The CRYSTAL experience. J Clin Oncol. 2008;26 Suppl:2. [Cited in This Article: ] |
| 9. | Bokemeyer C, Bondarenko I, Hartmann JT, De Braud FG, Volovat C, Nippgen J, Stroh C, Celik I, Koralewski P. KRAS status and efficacy of first-line treatment of patients with metastatic colorectal cancer (mCRC) with FOLFOX with or without cetuximab: The OPUS experience. J Clin Oncol. 2008;26 Suppl:4000. [Cited in This Article: ] |
| 10. | Tejpar S, Peeters M, Humblet Y, Vermorken JB, De Hertogh G, De Roock W, Nippgen J, von Heydebreck A, Stroh C, Van Cutsem E. Relationship of efficacy with KRAS status (wild type versus mutant) in patients with irinotecan-refractory metastatic colorectal cancer (mCRC), treated with irinotecan (q2w) and escalating doses of cetuximab (q1w): The EVEREST experience (preliminary data). J Clin Oncol. 2008;26 Suppl:4001. [Cited in This Article: ] |
| 11. | Jonker DJ, O'Callaghan CJ, Karapetis CS, Zalcberg JR, Tu D, Au HJ, Berry SR, Krahn M, Price T, Simes RJ. Cetuximab for the treatment of colorectal cancer. N Engl J Med. 2007;357:2040-2048. [Cited in This Article: ] |
| 12. | Wu CM, Tang R, Wang JY, Changchien CR, Hsieh LL. Frequency and spectrum of K-RAS codons 12 and 13 mutations in colorectal adenocarcinomas from Taiwan. Cancer Genet Cytogenet. 2005;158:55-60. [Cited in This Article: ] |
| 13. | Breivik J, Meling GI, Spurkland A, Rognum TO, Gaudernack G. K-ras mutation in colorectal cancer: relations to patient age, sex and tumour location. Br J Cancer. 1994;69:367-371. [Cited in This Article: ] |
| 14. | Kislitsin D, Lerner A, Rennert G, Lev Z. K-ras mutations in sporadic colorectal tumors in Israel: unusual high frequency of codon 13 mutations and evidence for nonhomogeneous representation of mutation subtypes. Dig Dis Sci. 2002;47:1073-1079. [Cited in This Article: ] |
| 15. | Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525-532. [Cited in This Article: ] |
| 16. | Zalis MG, Vieira FM, Zalcberg-Renault I, Bonamino MH, Ferreira CG, Oliveira S. KRAS mutation profile in colorectal cancer patients in Brazil: A cohort of 989 individuals. J Clin Oncol. 2009;27 Suppl:e15017. [Cited in This Article: ] |
| 17. | Poehlmann A, Kuester D, Meyer F, Lippert H, Roessner A, Schneider-Stock R. K-ras mutation detection in colorectal cancer using the Pyrosequencing technique. Pathol Res Pract. 2007;203:489-497. [Cited in This Article: ] |
| 18. | Ogino S, Kawasaki T, Brahmandam M, Yan L, Cantor M, Namgyal C, Mino-Kenudson M, Lauwers GY, Loda M, Fuchs CS. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7:413-421. [Cited in This Article: ] |
| 19. | Huncharek M, Muscat J, Geschwind JF. K-ras oncogene mutation as a prognostic marker in non-small cell lung cancer: a combined analysis of 881 cases. Carcinogenesis. 1999;20:1507-1510. [Cited in This Article: ] |