Abstract
Cell cycle progression is a process that is tightly controlled by internal and external signals. Environmental cues, such as those provided by growth factors, activate early signals that promote cell cycle entry1,2,3. Cells that have progressed past the restriction point become independent of growth factors, and cell cycle progression is then controlled endogenously. The phosphatidylinositol 3OH kinase (PI(3)K)/protein kinase B (PKB) pathway must be activated in G1 to inactivate forkhead transcription factors (FKH-TFs)4,5 and allow cell cycle entry2,3. Here we show that subsequent attenuation of the PI(3)K/PKB pathway is required to allow transcriptional activation of FKH-TF in G2. FKH-TF activity in G2 controls mammalian cell cycle termination, as interference with FKH transcriptional activation by disrupting PI(3)K/PKB downregulation, or by expressing a transcriptionally inactive FKH mutant, induces cell accumulation in G2/M, defective cytokinesis, and delayed transition from M to G1 of the cell cycle. We demonstrate that FKH-TFs regulate expression of mitotic genes such as cyclin B and polo-like kinase (Plk). Our results support the important role of forkhead in the control of mammalian cell cycle completion, and suggest that efficient execution of the mitotic programme depends on downregulation of PI(3)K/PKB and consequent induction of FKH transcriptional activity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Pardee, A. B. G1 events and regulation of cell proliferation. Science 246, 603–608 (1989).
Klippel, A. et al. Activation of phosphatidylinositol 3-kinase is sufficient for cell cycle entry and promotes cellular changes characteristic of oncogenic transformation. Mol. Cell. Biol. 18, 5699–5711 (1998).
Jones, S. M., Klinghoffer, R., Prestwich, G. D., Toker, A. & Kazlauskas, A. PDGF induces an early and a late wave of PI 3-kinase activity, and only the late wave is required for progression through G1. Curr. Biol. 9, 512–521 (1999).
Kops, G. J. et al. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature 398, 630–634 (1999).
Medema, R. H., Kops, G. J., Bos, J. L. & Burgering, B. M. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 404, 782–787 (2000).
Jiménez, C. et al. Identification and characterization of a new oncogene derived from the regulatory subunit of phosphoinositide 3-kinase. EMBO J. 17, 743–753 (1998).
Jiménez, C. et al. Role of the PI3K regulatory subunit in the control of actin organization and cell migration. J. Cell. Biol. 151, 249–262 (2000).
Tatsumoto, T., Xie, X., Blumenthal, R., Okamoto, I. & Miki, T. Human ECT2 is an exchange factor for Rho GTPases, phosphorylated in G2/M phases, and involved in cytokinesis. J. Cell. Biol. 147, 921–928 (1999).
Burgering, B. M. & Coffer, P. J. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature 376, 599–602 (1995).
del Peso, L., González, V. M., Hernández, R., Barr, F. G. & Núñez, G. Regulation of the forkhead transcription factor FKHR, but not the PAX3-FKHR fusion protein, by the serine/threonine kinase Akt. Oncogene 18, 7328–7333 (1999).
Brunet, A. et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96, 857–868 (1999).
Jackson, J. G., Kreisberg, J. I., Koterba, A. P., Yee, D. & Brattain, M. G. Phosphorylation and nuclear exclusion of the forkhead transcription factor FKHR after epidermal growth factor treatment in human breast cancer cells. Oncogene 19, 4574–4581 (2000).
Pic, A. et al. The forkhead protein Fkh2 is a component of the yeast cell cycle transcription factor SFF. EMBO J. 19, 3750–3761 (2000).
Zhu, G. et al. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature 406, 90–94 (2000).
Morgan, D. O. Regulation of the APC and the exit from mitosis. Nature Cell Biol. 1, E47–E53 (1999).
Murray, A. W., Solomon, M. J. & Kirschner, M. W. The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature 339, 280–286 (1989).
Surana, U. et al. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 12, 1969–1978 (1993).
Glover, D. M., Hagan, I. M. & Tavares, A. A. M. Polo-like kinases: a team that plays throughout mitosis. Genes Dev. 12, 3777–3787 (1998).
Kotani, S., Tanaka, H., Yasuda, H. & Todokoro, K. Regulation of APC activity by phosphorylation and regulatory factors. J. Cell. Biol. 146, 791–800 (1999).
Cogswell, J. P., Godlevski, M. M., Bonham, M., Bisi, J. & Babiss, L. Upstream stimulatory factor regulates expression of the cell cycle-dependent cyclin B1 promoter. Mol. Cell. Biol. 15, 2782–2790 (1995).
Bräuninger, A., Strebhardt, K. & Rübsamen-Waigmann, H. Identification and functional characterization of the human and murine polo-like kinase (Plk) promoter. Oncogene 11, 1793–1800 (1995).
Takeshita, T. et al. STAM, signal transducing adaptor molecule, is associated with Janus kinases and involved in signaling for cell growth and c-myc induction. Immunity 6, 449–457 (1997).
Carmena, M. et al. Drosophila polo kinase is required for cytokinesis. J. Cell. Biol. 143, 659–671 (1998).
Zachariae, W. Progression into and out of mitosis. Curr. Opin. Cell Biol. 11, 708–716 (1999).
Dijkers, P. F., Medema, R. H., Lammers, J. W., Koenderman, L. & Coffer, P. J. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr. Biol. 10, 1201–1204 (2000).
Borlado, L. R. et al. Increased phosphoinositide 3-kinase activity induces a lymphoproliferative disorder and contributes to tumor generation in vivo. FASEB J. 14, 895–903 (2000).
Reif, K., Nobes, C. D., Thomas, G., Hall, A. & Cantrell, D. A. Phosphatidylinositol 3-kinase signals activate a selective subset of Rac/Rho-dependent effector pathways. Curr. Biol. 6, 1445–1455 (1996).
Kotani, S. et al. PKA and MPF-activated polo-like kinase regulate anaphase-promoting complex activity and mitosis progression. Mol. Cell 1, 371–380 (1998).
Boyd, K. E., Wells, J., Gutman, J., Bartley, S. M. & Farnham, P. J. c-Myc target gene specificity is determined by a post-DNA-binding mechanism. Proc. Natl Acad. Sci. USA 95, 13887–13892 (1998).
Acknowledgements
We thank S. Gonzalo for advice in chromatin immunoprecipitation assays and M. Serrano, R. Medema, I. Mérida, M. Torres, D. Jones, V. Calvo and C. Mark for critical reading of the manuscript. This work was supported by grants from the Community of Madrid, DGCyT (Direccion General de Ciencia y Tecnologia) and the Pharmacia Corporation. The Department of Immunology and Oncology was founded and is supported by the Spanish Council for Scientific Research (CSIC) and by the Pharmacia Corporation.
Author information
Authors and Affiliations
Corresponding author
Supplementary information

Figure 1
(GIF 12.3 KB)
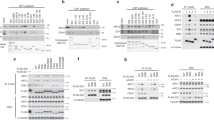
Constitutive but not transient activation of PI3K increases the percentage of cells in G2/M. (a) Expression of endogenous p85 (control) and recombinant inducible-p65PI3K in cells stably transfected with pRetroOn-p65PI3K. Induction was performed with doxycycline (2 mg/ml) for the indicated time periods and analysis was carried out by western blot using anti-p85 antibody. A representative assay of three with similar results is shown. (b) Cell lines stably expressing pRetroOn-p110CAAX (indicated as p110*) were incubated for 24 h in the presence (+) or absence (-) of doxycycline (2 mg/ml). Since anti-p110 Ab fail to detect p110 protein, cell extracts were examined by western blotting using anti-phospho-Ser473-PKB antibody (upper) or anti-PKB antibody (lower), revealing the activation of PKB by p110CAAX. (c) % of cells in G2/M of one representative cell line expressing pRetroOn-p65PI3K and two representative cell lines expressing RetroOn-p110CAAX. Cells were incubated for 48 h in medium with serum in the presence or absence of doxycycline (2 mg/ml) and then examined.

Figure 1
(JPG 54.3 KB)
Mitotic staining of control cells and cells expressing a constitutive active form of PI3K. Exponentially growing control NIH-3T3 cells, or NIH-3T3 cells expressing p110CAAX (p110*) were stained with an anti-tubulin antibody and anti-rabbit-Cy3 (red), FITC/phalloidin (green) and Hoechst 33258 (blue) for simultaneous visualization of the mitotic spindle, the actomyosin contractile ring and chromatin, respectively as described (Tatsumoto, T., et al., J Cell Biol 147, 921, 1999).
Supplementary Information Methods
Luciferase reporter constructs and oligonucleotides for ChIP assays
We identified two regions in the human plk and cyclin B promoters containing sequences similar to FKH binding sites (Fig. 4c). DNA fragments including these regions were obtained from HeLa total genomic DNA by PCR. These fragments were inserted in the polylinker of the pGL3-promoter vector (Promega), upstream of the SV40 minimal promoter controlling luciferase expression. PCR reactions were also performed following chromatin immunoprecipitation (ChIP assays). The following oligonucleotides (some of which were designed to include a restriction site) were used:
For cyclin B region I-Luc, the 5´ oligonucleotide at position –542 of the cyclin B promoter was GCTGCTACCGTAGAAATGGAAAG; the 3´ oligonucleotide at position –254 of the cyclin B promoter was GATCGCGCGAAGGCGTTCGA. The PCR fragment was cleaved using the BsmAI restriction enzyme, treated with Klenow, and cleaved with XmnI. The fragment was subcloned into pGL3 cleaved with SmaI. The same oligonucleotides were used for cyclin B-region I PCR in ChIP assays.
For cyclin B region II-Luc, the 5´ oligonucleotide at position –1015 of the cyclin B promoter was TTGGATCTAGAGAGAATCTGAGC; the 3´ oligonucleotide at position –786 of the promoter was ACTTGATTACCTCGAGATTGTCC. The PCR fragment was digested using XbaI (NheI-compatible) and XhoI restriction enzymes, and was subcloned into pGL3, previously cleaved with NheI and XhoI. For ChIP assays, another fragment including the cyclin B-region II was amplified by PCR using the 5´ oligonucleotide at position –949, TTCCTGATTTTCCCATGAGAGGC, and the 3´ oligonucleotide at position –657, GCCAGGGTCACACATTAGCAAC. Although the first oligonucleotide combination worked well in ChIP assays, the second combination (at -949 and -657) yielded optimal results.
For plk region I-Luc, the 5´ oligonucleotide at position –786 of the plk promoter was TCCAGCCCGGGCGACAG; the 3´ oligonucleotide at position –353 was CGGGAAAGATCTGCGGTTCAC. The PCR fragment was cleaved with MaeIII, treated with Klenow, and then cleaved with BglII. This fragment was inserted into pGL3, previously digested with SmaI and Bgl II. The same oligonucleotides were used in ChIP assays´ PCRs.
For plk region II-Luc, the 5´ oligonucleotide at position –2403 of the plk promoter was CTGCAGTGAGCCAAGATCAC; the 3´ oligonucleotide at position -1843 was TCCTCCAGCAATCAGTTGTGAC. The PCR fragment was cleaved with MaeIII, Klenow-treated and inserted into SmaI-digested pGL3. For ChIP assays, a smaller fragment was amplified using the 5´ oligonucleotide at position –2362, GTGACAGAGCAAGA CTCCATC, and the 3´ oligonucleotide at position –1980, AAGCAGATGTTGGGCAT AACC.
Following ChIP assays, a region of the human cyclin D3 promoter (negative control) and the human Fas ligand promoter (positive control) were also amplified by PCR using the 5´ oligonucleotide at position –901 of the cyclin D3 promoter, GCAACTTAACAAGGTG AGGCC, and the 3´ oligonucleotide at position –589 of the promoter ATGACTCACTA CATCTTCACGAA. For the Fas ligand promoter, the 5´ oligonucleotide at position -1057 was TAGTCAGGTGTAGTGACTTATGC, and the 3´ oligonucleotide at position -756 was CTGACCTGCCGATCACCATAAT
Rights and permissions
About this article
Cite this article
Alvarez, B., Martínez-A., C., Burgering, B. et al. Forkhead transcription factors contribute to execution of the mitotic programme in mammals. Nature 413, 744–747 (2001). https://doi.org/10.1038/35099574
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35099574
This article is cited by
-
Public RNA-seq data-based identification and functional analyses reveal that MXRA5 retains proliferative and migratory abilities of dental pulp stem cells
Scientific Reports (2023)
-
Targeted inhibition of metastatic melanoma through interference with Pin1-FOXM1 signaling
Oncogene (2016)
-
FOXC2 regulates the G2/M transition of stem cell-rich breast cancer cells and sensitizes them to PLK1 inhibition
Scientific Reports (2016)
-
FoxO3 suppresses Myc-driven lymphomagenesis
Cell Death & Disease (2016)
-
Forkhead box K2 modulates epirubicin and paclitaxel sensitivity through FOXO3a in breast cancer
Oncogenesis (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.