Abstract
Background:
We aimed to compare the sensitive and quality-controlled KRAS testing with direct sequencing and to assess the impact on decision making of treatment.
Patients and methods:
We analysed genomic DNA isolated from macrodissected formalin-fixed paraffin-embedded specimens by direct sequencing and an amplification refractory mutation system–Scorpion assay (ARMS/S) method. Cetuximab was administered to patients identified as having wild-type (WT) KRAS using direct sequencing. Therapeutic effects were evaluated according to their KRAS status as determined by ARMS/S.
Results:
Among the 159 patients, the overall mutation rate was determined to be 37.0% by direct sequencing and 44.0% by ARMS/S. For the patients diagnosed as WT by direct sequencing and treated with cetuximab (n=47), a response rate of 16.0% was observed for 38 ARMS/S WT patients, whereas 9 ARMS/S mutant (MUT) patients failed to respond. The ARMS/S WT patients showed significant improvement in progression-free survival (PFS) and overall survival (OS) compared with ARMS/S MUT patients (PFS median 5.0 vs 1.7 months, hazards ratio (HR)=0.29, P=0.001; OS median 12.1 vs 3.8 months, HR=0.26, P=0.001).
Conclusion:
Sensitive and quality-controlled KRAS testing may provide improved predictive power to determine the efficacy of anti-epidermal growth factor antibodies.
Similar content being viewed by others
Main
Retrospective subset analyses and prospective randomised phase III clinical trials have suggested that anti-epidermal growth factor antibodies do not benefit patients with metastatic colorectal cancer harbouring KRAS mutations (Amado et al, 2008; Karapetis et al, 2008; Tol et al, 2009; Van Cutsem et al, 2009). On the basis of these findings, regulatory authorities in Europe, the United States and Japan require pretreatment KRAS mutation testing. In Europe, the KRAS European Quality Assurance Program (http://kras.eqascheme.org/) has been launched and several Communauté Européene-labelled KRAS mutation test kits such as the TheraScreen K-RAS Mutation Kit (DxS-QIAGEN, Manchester, UK), KRAS LightMix (TIB MolBiol, Berlin, Germany) and PyroMark Q24 KRAS Kit (QIAGEN, Duesseldorf, Germany) have been approved for diagnostic use. The TheraScreen Kit combines the amplification refractory mutation system (ARMS) with a unique bifunctional fluorescent primer/probe molecule (Scorpion) and is recommended for clinical use because of its high sensitivity, robustness and convenience (Franklin et al, 2009; Jimeno et al, 2009; Kotoula et al, 2009; Whitehall et al, 2009; Angulo et al, 2010; Ogasawara et al, 2011).
Together with these standardised methods, direct sequencing is still one of the most accessible methods. However, several critical disadvantages of direct sequencing for diagnostic use have been indicated. These include its low sensitivity and lack of strict criteria for distinguishing mutant signals from contaminated noises. Furthermore, we have recently reported that insufficient PCR amplification further limits the sensitivity and specificity of direct sequencing. This is particularly important when DNA isolated from formalin-fixed paraffin-embedded (FFPE) specimens, which contain low levels of amplifiable DNA, is used (Bando et al, 2011). To increase the sensitivity of direct sequencing, macroscopic isolation of tissues in which cancer cells occupy >70% of the area (macrodissection) is recommended for preparation of genomic DNA (Kotoula et al, 2009).
Although discrepancies in interpretation between the ARMS–Scorpion assay (ARMS/S) and direct sequencing have been noted, the impact of these discrepancies on treatment has not been adequately evaluated (Franklin et al, 2009; Kotoula et al, 2009). In this study, we evaluated the validity of ARMS/S and direct sequencing by comparing the therapeutic effects of cetuximab in patients in whom KRAS mutations were analysed by these two methods.
Patients and methods
DNA samples and KRAS mutation testing
Genomic DNA was extracted from primary and metastatic colorectal cancer tissues of patients scheduled to receive cetuximab. DNA extraction from FFPE tissue blocks has been previously described. The KRAS exon-2 fragment was amplified and sequenced according to previously described methods (Bando et al, 2011). The KRAS PCR Kit (DxS-QIAGEN) was used for detection of seven major mutations in KRAS codons 12 and 13. Reactions were performed using the LightCycler 480 Real-Time PCR System (Roche Diagnostics, Mannheim, Germany) and analysed with LightCycler Adapt software v1.1 (Roche Diagnostics) as previously described (Bando et al, 2011).
Patients
Cetuximab was administered at the National Cancer Center Hospital East (NCCHE) in patients diagnosed with wild-type (WT) KRAS by direct sequencing. Furthermore, KRAS mutation status was evaluated using ARMS/S.
Patients who met all inclusion criteria were retrospectively included in analyses. Inclusion criteria were as follows: (1) age ⩾20 years; (2) histologically confirmed adenocarcinoma of the colon or rectum; (3) presence of unresectable metastatic disease; (4) baseline computed tomography (CT) scan performed within the previous 28 days; (5) initial evaluation by CT scan within 3 months; (6) documentation of refractory to previous fluoropyrimidine, oxaliplatin and irinotecan administration; (7) KRAS mutational status determined by direct sequencing and ARMS/S; (8) Eastern Cooperative Oncology Group performance status score ⩽2; (9) adequate haematological, hepatic, renal and bone marrow function; and (10) undergone treatment with cetuximab monotherapy regimen or combination regimen with cetuximab plus irinotecan. In the monotherapy regimen, cetuximab was administered at an initial dose of 400 mg m–2, followed by weekly infusions of 250 mg m–2. In the combination regimen, cetuximab was administered at the same dose as for monotherapy, followed by biweekly infusions of 150 mg m–2 irinotecan.
The study was conducted with the approval of the institutional review board.
Measured outcomes
The therapeutic response rate was evaluated according to the Response Evaluation Criteria in Solid Tumours (version 1.0). Progression-free survival (PFS) was defined as the time from the first cetuximab administration to either first objective evidence of disease progression or death from any cause. Overall survival (OS) was defined as the time from the first administration of cetuximab to death from any cause.
Statistical analysis
The response rate, PFS and OS of all patients were revalued for this study. Fisher's exact test and the Mann–Whitney test were used to compare the patient characteristics and response rates. The PFS and OS data were plotted as Kaplan–Meier curves and the differences between the groups categorised by ARMS/S-identified KRAS status were compared by the log-rank test. The hazard ratio (HR) was calculated from the Cox regression model with a single covariate. All analyses were performed using IBM SPSS Statistics 18 package software (SPSS Inc., Tokyo, Japan).
Results
Mutation rates determined by direct sequencing and ARMS/S
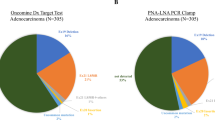
From April 2009 to March 2010, 159 specimens were tested using both ARMS/S and direct sequencing (98 specimens were collected from NCCHE and 61 from other hospitals). Both methods had a success rate of 100%. In all, 59 (37.0%) KRAS mutations were detected by direct sequencing and 70 (44.0%) by ARMS/S (Table 1a). All mutations identified by direct sequencing were also identified by ARMS/S. However, 11 (7.0%) of the 70 KRAS mutations identified by ARMS/S were not detected by direct sequencing. The overall concordance rate of the two methods was 93.0% (Table 1b).
Patient characteristics
From April 2009 to March 2010, 47 patients met with all of the inclusion criteria (11 patients were treated with cetuximab monotherapy and 36 patients were treated with cetuximab plus irinotecan). Of the 47 patients, 38 and 9 patients were identified by ARMS/S as WT (ARMS/S WT) and mutant (ARMS/S MUT), respectively (Table 2). Patient characteristics of the two groups (ARMS/S WT vs ARMS/S MUT) were not significantly different except for the incidence of lung metastases (Table 2).
Response to treatment
The response rate of ARMS/S WT patients was 16.0%. In contrast, no objective tumour response was observed in ARMS/S MUT patients. In addition, the disease control rates (including partial response and stable disease) of ARMS/S WT and ARMS/S MUT patients were 66.0% and 56.0%, respectively (Table 3).
Survival
The median PFS of the 38 ARMS/S WT and 9 ARMS/S MUT patients was 5.0 and 1.7 months, respectively (HR=0.29, P=0.001; Table 3 and Figure 1A). The relative dose intensity of cetuximab therapy was not significantly different between ARMS/S WT and ARMS/S MUT patients (Table 3). The median OS of the 38 ARMS/S WT and 9 ARMS/S MUT patients was 12.1 and 3.8 months, respectively (HR=0.26, P=0.001; Table 3 and Figure 1B).
(A) Kaplan–Meier plots of progression-free survival (PFS) according to KRAS status determined by the amplification refractory mutation system–Scorpion assay (ARMS/S). For the patients treated with cetuximab-containing regimens, the median PFS values were 5.0 and 1.7 months for ARMS/S wild-type (solid line) and ARMS/S mutant (dashed line) patients, respectively. The difference was statistically significant (HR=0.29, P=0.001). (B) Kaplan–Meier plots of overall survival (OS) according to KRAS status determined by ARMS/S. For the patients treated with cetuximab-containing regimens, the median OS values for ARMS/S wild-type (solid line) and ARMS/S mutant (dashed line) patients were 12.1 and 3.8 months, respectively. The difference was statistically significant (HR=0.26, P=0.001).
When the patients were divided as per treatment regimen, the median PFS and OS of the patients treated with cetuximab plus irinotecan were significantly different from ARMS/S WT and ARMS/S MUT patients (PFS; HR=0.23, P=0.002, OS; HR=0.187, P=0.001). Similar trends were also observed for the patients treated with cetuximab monotherapy (PFS; HR=0.497, P=0.332, OS; HR=0.674, P=0.586).
Discussion
The present guidelines for KRAS testing allow direct sequencing for MUT detection (Allegra et al, 2009). To overcome the low sensitivity of direct sequencing, we performed macrodissection of the tissues in order to enrich the tumour cell-derived DNA. We also improved the PCR conditions based on our previous study (Bando et al, 2011). The mutation rates determined by direct sequencing in the present study were comparable with those reported in previous studies (36.0–43.0%) as per various mutation detection methods, including ARMS/S, and thus support the validity of our direct sequencing procedure (Amado et al, 2008; Karapetis et al, 2008; Tol et al, 2009; Van Cutsem et al, 2009). In contrast, the mutation rate determined using simultaneous ARMS/S appeared to be higher than that found in previous clinical trials (Amado et al, 2008; Karapetis et al, 2008; Tol et al, 2009; Van Cutsem et al, 2009). Therefore, we surmise that enrichment of tumour cell-derived DNA may further enhance the sensitivity of ARMS/S.
Next, we examined whether this higher sensitivity could result in improved clinical relevance. The median PFS, OS and response rates of KRAS WT patients determined by ARMS/S were comparable with those previously reported (median PFS, 3.8 months; median OS, 9.5 months; response rate, 13.0%) (Karapetis et al, 2008). In contrast, the median PFS, OS and response rates of the KRAS MUT patients, although determined as WT by direct sequencing, were comparable with those of MUT KRAS patients reported in previous clinical trials (median PFS, 1.8 months; median OS, 4.5 months; response rate, 1.0%) (Karapetis et al, 2008).
Two factors may be responsible for the significant advantage of ARMS/S. First, the higher sensitivity of the ARM/S assay can detect the presence of a lesser number of KRAS mutations than direct sequencing. Second, strictly controlled criteria for MUT identification provided robust detection and eliminated the ‘grey zone’ cases that we often encountered using direct sequencing.
On the other hand, the intratumoral heterogeneity of tumour tissues for KRAS gene status suggested that residual KRAS WT tumour cells may respond to cetuximab, but this idea is still under debate (Baldus et al, 2010). In the present study, although ARMS/S MUT patients showed poorer PFS and OS than the WT patients, 5 of the 9 patients achieved disease stability in the first CT evaluation. Although this study had limitations such as small sample size and retrospective design that could have caused substantial biases, it appears that tumour heterogeneity allowed a reasonable level of disease control. Thus, further evaluation with an adequate sample size, in a prospective manner, would be required to determine which of the testing methods (direct sequencing or ARMS/S) would be a better predictive marker for clinical benefits.
In conclusion, our study suggested that KRAS mutation status determined by ARMS/S appeared to be more closely related to clinical effects than that determined by direct sequencing, although there were limitations of sample size and retrospective design. Whether KRAS mutation status determined by ARMS/S can be used as a predictive biomarker is not yet known. However, the study results warrant further investigation of this method, which should evaluate the correlations between KRAS mutation status and clinical outcomes in comparison with those achieved by direct sequencing.
Change history
29 March 2012
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, McAllister PK, Morton RF, Schilsky RL (2009) American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol 27: 2091–2096
Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD (2008) Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 26: 1626–1634
Angulo B, Garcia-Garcia E, Martinez R, Suarez-Gauthier A, Conde E, Hidalgo M, Lopez-Rios F (2010) A commercial real-time PCR kit provides greater sensitivity than direct sequencing to detect KRAS mutations: a morphology-based approach in colorectal carcinoma. J Mol Diagn 12: 292–299
Baldus SE, Schaefer KL, Engers R, Hartleb D, Stoecklein NH, Gabbert HE (2010) Prevalence and heterogeneity of KRAS, BRAF and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res 16: 790–799
Bando H, Tsuchihara K, Yoshino T, Kojima M, Ogasawara N, Fukushima H, Ochiai A, Ohtsu A, Esumi H (2011) Biased discordance of KRAS mutation detection in archived colorectal cancer specimens between the ARMS-Scorpion method and direct sequencing. Jpn J Clin Oncol 41: 239–244
Franklin WA, Haney J, Sugita M, Bemis L, Jimeno A, Messersmith WA (2009) KRAS mutation: comparison of testing methods and tissue sampling techniques in colon cancer. J Mol Diagn 12: 43–50
Jimeno A, Messersmith WA, Hirsch FR, Franklin WA, Eckhardt SG (2009) KRAS mutations and sensitivity to epidermal growth factor receptor inhibitors in colorectal cancer: practical application of patient selection. J Clin Oncol 27: 1130–1136
Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au HJ, Langer C, Moore MJ, Zalcberg JR (2008) K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 359: 1757–1765
Kotoula V, Charalambous E, Biesmans B, Malousi A, Vrettou E, Fountzilas G, Karkavelas G (2009) Targeted KRAS mutation assessment on patient tumour histologic material in real time diagnostics. PLoS ONE 4: e7746
Ogasawara N, Bando H, Kawamoto Y, Yoshino T, Tsuchihara K, Ohtsu A, Esumi H (2011) Feasibility and robustness of amplification refractory mutation system (ARMS)-based KRAS testing using clinically available formalin-fixed, paraffin-embedded samples of colorectal cancers. Jpn J Clin Oncol 41: 52–56
Tol J, Koopman M, Cats A, Rodenburg CJ, Creemers GJ, Schrama JG, Erdkamp FL, Vos AH, van Groeningen CJ, Sinnige HA, Richel DJ, Voest EE, Dijkstra JR, Vink-Borger ME, Antonini NF, Mol L, van Krieken JH, Dalesio O, Punt CJ (2009) Chemotherapy, bevacizumab and cetuximab in metastatic colorectal cancer. N Engl J Med 360: 563–572
Van Cutsem E, Kohne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D’Haens G, Pinter T, Lim R, Bodoky G, Roh JK, Folprecht G, Ruff P, Stroh C, Tejpar S, Schlichting M, Nippgen J, Rougier P (2009) Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med 360: 1408–1417
Whitehall V, Tran K, Umapathy A, Grieu F, Hewitt C, Evans TJ, Ismail T, Li WQ, Collins P, Ravetto P, Leggett B, Salto-Tellez M, Soong R, Fox S, Scott RJ, Dobrovic A, Iacopetta B (2009) A multicenter blinded study to evaluate KRAS mutation testing methodologies in the clinical setting. J Mol Diagn 11: 543–552
Acknowledgements
This work was partly supported by ‘Grants for the Third-Term Comprehensive 10-Year Strategy for Cancer Control’ and ‘Grant-in-Aid for Cancer Research from the Ministry of Health, Labor and Welfare, Japan’.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict interest.
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Bando, H., Yoshino, T., Tsuchihara, K. et al. KRAS mutations detected by the amplification refractory mutation system–Scorpion assays strongly correlate with therapeutic effect of cetuximab. Br J Cancer 105, 403–406 (2011). https://doi.org/10.1038/bjc.2011.247
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2011.247
Keywords
This article is cited by
-
Comparative sequence analysis of patient-matched primary colorectal cancer, metastatic, and recurrent metastatic tumors after adjuvant FOLFOX chemotherapy
BMC Cancer (2019)
-
Enrichment technique to allow early detection and monitor emergence of KRAS mutation in response to treatment
Scientific Reports (2019)
-
Improved detection of BRAF V600E using allele-specific PCR coupled with external and internal controllers
Scientific Reports (2017)
-
Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA
Nature Medicine (2014)
-
Simultaneous identification of 36 mutations in KRAS codons 61and 146, BRAF, NRAS, and PIK3CAin a single reaction by multiplex assay kit
BMC Cancer (2013)