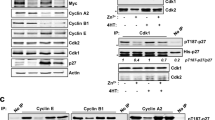
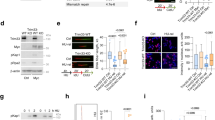
Abstract
The c-Myc proto-oncogene encodes a transcription factor that is essential for cell growth and proliferation and is broadly implicated in tumorigenesis. However, the biological functions required by c-Myc to induce oncogenesis remain elusive. Here we show that c-Myc has a direct role in the control of DNA replication. c-Myc interacts with the pre-replicative complex and localizes to early sites of DNA synthesis. Depletion of c-Myc from mammalian (human and mouse) cells as well as from Xenopus cell-free extracts, which are devoid of RNA transcription, demonstrates a non-transcriptional role for c-Myc in the initiation of DNA replication. Overexpression of c-Myc causes increased replication origin activity with subsequent DNA damage and checkpoint activation. These findings identify a critical function of c-Myc in DNA replication and suggest a novel mechanism for its normal and oncogenic functions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
29 June 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41586-023-06284-1
References
Dalla-Favera, R. et al. Cloning and characterization of different human sequences related to the onc gene (v-myc) of avian myelocytomatosis virus (MC29). Proc. Natl Acad. Sci. USA 79, 6497–6501 (1982)
Vennstrom, B., Sheiness, D., Zabielski, J. & Bishop, J. M. Isolation and characterization of c-myc, a cellular homolog of the oncogene (v-myc) of avian myelocytomatosis virus strain 29. J. Virol. 42, 773–779 (1982)
Grandori, C., Cowley, S. M., James, L. P. & Eisenman, R. N. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16, 653–699 (2000)
Fernandez, P. C. et al. Genomic targets of the human c-Myc protein. Genes Dev. 17, 1115–1129 (2003)
Patel, J. H., Loboda, A. P., Showe, M. K., Showe, L. C. & McMahon, S. B. Analysis of genomic targets reveals complex functions of MYC. Nature Rev. Cancer 4, 562–568 (2004)
Dalla-Favera, R. et al. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc. Natl Acad. Sci. USA 79, 7824–7827 (1982)
Dalla-Favera, R., Wong-Staal, F. & Gallo, R. C. Onc gene amplification in promyelocytic leukaemia cell line HL-60 and primary leukaemic cells of the same patient. Nature 299, 61–63 (1982)
Pasqualucci, L. et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature 412, 341–346 (2001)
Collins, S. & Groudine, M. Amplification of endogenous myc-related DNA sequences in a human myeloid leukaemia cell line. Nature 298, 679–681 (1982)
Adams, J. M. et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 318, 533–538 (1985)
Pelengaris, S., Khan, M. & Evan, G. c-MYC: more than just a matter of life and death. Nature Rev. Cancer 2, 764–776 (2002)
Wade, M. & Wahl, G. M. c-Myc, genome instability, and tumorigenesis: the devil is in the details. Curr. Top. Microbiol. Immunol. 302, 169–203 (2006)
Machida, Y. J., Hamlin, J. L. & Dutta, A. Right place, right time, and only once: replication initiation in metazoans. Cell 123, 13–24 (2005)
Gilbert, D. M. In search of the holy replicator. Nature Rev. Mol. Cell Biol. 5, 848–855 (2004)
Iguchi-Ariga, S. M., Itani, T., Kiji, Y. & Ariga, H. Possible function of the c-myc product: promotion of cellular DNA replication. EMBO J. 6, 2365–2371 (1987)
Obaya, A. J., Mateyak, M. K. & Sedivy, J. M. Mysterious liaisons: the relationship between c-Myc and the cell cycle. Oncogene 18, 2934–2941 (1999)
Koch, H. B. et al. Large-scale identification of c-MYC-associated proteins using a combined TAP/MudPIT approach. Cell Cycle 6, 205–217 (2007)
McMahon, S. B., Van Buskirk, H. A., Dugan, K. A., Copeland, T. D. & Cole, M. D. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94, 363–374 (1998)
Zhang, J. J. et al. Ser727-dependent recruitment of MCM5 by Stat1α in IFN-γ-induced transcriptional activation. EMBO J. 17, 6963–6971 (1998)
Facchini, L. M., Chen, S., Marhin, W. W., Lear, J. N. & Penn, L. Z. The Myc negative autoregulation mechanism requires Myc-Max association and involves the c-myc P2 minimal promoter. Mol. Cell. Biol. 17, 100–114 (1997)
Grignani, F. et al. Negative autoregulation of c-myc gene expression is inactivated in transformed cells. EMBO J. 9, 3913–3922 (1990)
Abdurashidova, G. et al. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 287, 2023–2026 (2000)
Tao, L., Dong, Z., Leffak, M., Zannis-Hadjopoulos, M. & Price, G. Major DNA replication initiation sites in the c-myc locus in human cells. J. Cell. Biochem. 78, 442–457 (2000)
Prathapam, T., Tegen, S., Oskarsson, T., Trumpp, A. & Martin, G. S. Activated Src abrogates the Myc requirement for the G0/G1 transition but not for the G1/S transition. Proc. Natl Acad. Sci. USA 103, 2695–2700 (2006)
Rao, P. N. & Johnson, R. T. Mammalian cell fusion: studies on the regulation of DNA synthesis and mitosis. Nature 225, 159–164 (1970)
Blow, J. J. & Laskey, R. A. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell 47, 577–587 (1986)
Walter, J., Sun, L. & Newport, J. Regulated chromosomal DNA replication in the absence of a nucleus. Mol. Cell 1, 519–529 (1998)
Grandori, C. et al. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nature Cell Biol. 7, 311–318 (2005)
Woodward, A. M. et al. Excess Mcm2–7 license dormant origins of replication that can be used under conditions of replicative stress. J. Cell Biol. 173, 673–683 (2006)
Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S. & Bonner, W. M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 (1998)
Costanzo, V. et al. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol. Cell 8, 137–147 (2001)
McGarry, T. J. & Kirschner, M. W. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93, 1043–1053 (1998)
Wang, X. et al. Role of DNA replication proteins in double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 24, 6891–6899 (2004)
Shechter, D., Costanzo, V. & Gautier, J. ATR and ATM regulate the timing of DNA replication origin firing. Nature Cell Biol. 6, 648–655 (2004)
Kovalchuk, A. L. et al. Burkitt lymphoma in the mouse. J. Exp. Med. 192, 1183–1190 (2000)
Knoepfler, P. S. et al. Myc influences global chromatin structure. EMBO J. 25, 2723–2734 (2006)
Norio, P. DNA replication: the unbearable lightness of origins. EMBO Rep. 7, 779–781 (2006)
Bartkova, J. et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434, 864–870 (2005)
Gorgoulis, V. G. et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434, 907–913 (2005)
Felsher, D. W. & Bishop, J. M. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc. Natl Acad. Sci. USA 96, 3940–3944 (1999)
Mai, S. & Mushinski, J. F. c-Myc-induced genomic instability. J. Environ. Pathol. Toxicol. Oncol. 22, 179–199 (2003)
Grandori, C. et al. Werner syndrome protein limits MYC-induced cellular senescence. Genes Dev. 17, 1569–1574 (2003)
Cobb, J. A. et al. Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev. 19, 3055–3069 (2005)
Di Micco, R. et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444, 638–642 (2006)
Felsher, D. W., Zetterberg, A., Zhu, J., Tlsty, T. & Bishop, J. M. Overexpression of MYC causes p53-dependent G2 arrest of normal fibroblasts. Proc. Natl Acad. Sci. USA 97, 10544–10548 (2000)
McCormack, S. J. et al. Myc/p53 interactions in transgenic mouse mammary development, tumorigenesis and chromosomal instability. Oncogene 16, 2755–2766 (1998)
Nikolaev, A. Y. Li, M. Puskas, N., Qin, J. & Gu, W. Parc: a cytoplasmic anchor for p53. Cell 112, 29–40 (2003)
Grandori, C. et al. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nature Cell Biol. 7, 311–318 (2005)
Smythe, C. &. Newport, J. W. Systems for the study of nuclear assembly, DNA replication, and nuclear breakdown in Xenopus laevis egg extracts. Methods Cell Biol. 35, 449–468 (1991)
Walter, J. Sun, L. & Newport, J. Regulated chromosomal DNA replication in the absence of a nucleus. Mol Cell 1, 519–529 (1998)
Michael, W. M. Ott, R. Fanning, E. & Newport, J. Activation of the DNA replication checkpoint through RNA synthesis by primase. Science 289, 2133–2137 (2000)
Hickson, I. et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res 64, 9152–9159 (2004)
Jackson, J. R. et al. An indolocarbazole inhibitor of human checkpoint kinase (Chk1) abrogates cell cycle arrest caused by DNA damage. Cancer Res 60, 566–572 (2000)
Kovalchuk, A. L. et al. Burkitt lymphoma in the mouse. J. Exp. Med. 192, 1183–1190 (2000)
Acknowledgements
We thank R. Baer for critical reading of the manuscript; J. Walter and H. Nishitani for Xenopus Cdc45 and human Cdt1 antibodies; W. M. Michael for DNA polymerase α antibodies; D. Shechter for NPE and technical suggestions; A. Lasorella and A. Iavarone for N-Myc plasmids; G. S. Martin and T. Prathapam for 3T9 c-myc+/- cells; W. Zhang for mass spectrometry analysis; P. Liccardo for generating the Myc H1299 cell line; G. Cattoreti, P. Smith and J. Kosek for technical advice with the immunofluorescence; H. Morse for providing the λ-Myc mouse strain; U. Klein and L. Pasqualucci for advice on mouse B-cell isolation; C. Li for Myc siRNA sequences; M. Lia for GAPDH primers; C. Franci for advice on peptide selection for XMyc antibody; and K. Robinson for technical help with ChIP, quantitative PCR and deconvolution microscopy analysis. This work was supported by grants from the National Institutes of Health (to R.D.-F. and J.G.) and the American Cancer Society (to J.G.). D.D.-S. was a recipient of a postdoctoral grant from Fundacio “la Caixa” (Spain).
Author Contributions R.D.-F. and J.G. supervised the entire project and contributed equally as co-senior authors; D.D.-S. designed and conducted the experiments in mammalian cells; D.D.-S. and C.Y.Y. designed the experiments in Xenopus, which were conducted by C.Y.Y.; D.D.-S. and C.Y.Y. wrote the manuscript under the supervision of R.D.-F. and J.G. and comments from all co-authors; C.G. designed and conducted immunofluorescence and ChIP experiments in human primary cells; B.C. and M.L. conducted the initial purification and chromatography of the Myc complex, designed and supervised by W.G.; and L.R. contributed to the biochemical characterization of the complex.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Information 1
This file contains Supplementary Figures 1-11 with Legends, Supplementary Table 1 Supplementary Methods and Legends to the Supplementary Videos 1-3. (PDF 1858 kb)
Supplementary Video 1
This file contains Supplementary Video 1 (MOV 818 kb)
Supplementary Video 2
This file contains Supplementary Video 2 (MOV 3194 kb)
Supplementary Video 3
This file contains Supplementary Video 3 (MOV 3238 kb)
Rights and permissions
About this article
Cite this article
Dominguez-Sola, D., Ying, C., Grandori, C. et al. Non-transcriptional control of DNA replication by c-Myc. Nature 448, 445–451 (2007). https://doi.org/10.1038/nature05953
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature05953
This article is cited by
-
Oncogenic c-Myc induces replication stress by increasing cohesins chromatin occupancy in a CTCF-dependent manner
Nature Communications (2024)
-
WWOX promotes osteosarcoma development via upregulation of Myc
Cell Death & Disease (2024)
-
cGAS-STING pathway expression correlates with genomic instability and immune cell infiltration in breast cancer
npj Breast Cancer (2024)
-
NFIB facilitates replication licensing by acting as a genome organizer
Nature Communications (2023)
-
In vitro antiproliferative activity of Parrotia persica exclusive gallotannin
Medicinal Chemistry Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.