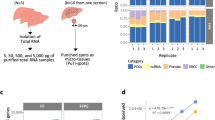
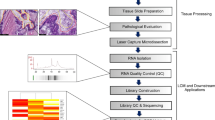
Abstract
Breast cancer is a heterogenous disease in terms of both clinical behavior and molecular characteristics. To develop prognostic and predictive markers for breast cancer, it would be useful to be able to analyze formalin-fixed paraffin-embedded tissue (FPET) collected and banked from completed clinical trials. RNAs extracted from FPETs are chemically modified and fragmented, and are therefore not ideal substrates for gene-expression profiling assays. However, methods are being developed to optimize the use of such RNAs for high-throughput gene expression profiling assays. For microarray analysis, existing methods may be adequate for fresh FPET, but they do not work well with older FPET. For older samples, real-time reverse transcription-polymerase chain reaction is the method of choice for gene-expression profiling.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Volpi A et al. (2004) Prognostic relevance of histological grade and its components in node-negative breast cancer patients. Mod Pathol 17: 1038–1044
Schena M et al. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270: 467–470
Perou CM et al. (2000) Molecular portraits of human breast tumours. Nature 406: 747–752
van't Veer LJ et al. (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415: 530–536
van de Vijver MJ et al. (2002) A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347: 1999–2009
Sorlie T et al. (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 98: 10869–10874
Masuda N et al. (1999) Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res 27: 4436–4443
Cronin M et al. (2004) Measurement of gene expression in archival paraffin-embedded tissues: development and performance of a 92-gene reverse transcriptase-polymerase chain reaction assay. Am J Pathol 164: 35–42
Affymetrix® Genechip Arrays—GeneChip® Human X3P Array [http://www.affymetrix.com/products/arrays/specific/x3p.affx]
Applied Biosystems Genomic Products TaqMan® gene expression assay
Dheda K et al. (2004) Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 37: 112–119
Ma XJ et al. (2004) A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell 5: 607–616
Genomic Health™ Oncotype DX™ Breast Cancer Assay [http://www.genomichealth.com/oncotype/default.aspx]
Paik S et al. (2004) A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 351: 2817–2826
Collins LC et al. (2005) Bimodal frequency distribution of estrogen receptor immunohistochemical staining results in breast cancer: an analysis of 825 cases. Am J Clin Pathol. 123: 16–20
Mikhitarian K et al. (2004) Enhanced detection of RNA from paraffin-embedded tissue using a panel of truncated gene-specific primers for reverse transcription. Biotechniques 36: 474–478
Illumina® DASL™ assay [http://www.illumina.com/products/geneexpression/dasl_assay.ilmn]
Bibikova M et al. (2004) Quantitative gene expression profiling in formalin-fixed, paraffin-embedded tissues using universal bead arrays. Am J Pathol 165: 1799–1807
Pollack JR et al. (2002) Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci U S A 99: 12963–12968
Hyman E et al. (2002) Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res 62: 6240–6245
Roche PC et al. (2002) Concordance between local and central laboratory HER2 testing in the breast intergroup trial N9831. J Natl Cancer Inst 94: 855–857
Acknowledgements
The studies mentioned in this article were in part funded by the following grants: NCI grant U10CA12027 awarded to NSABP Foundation Inc (Pittsburgh, PA), Common Wealth Universal Research Enhancement Program from PA State Department of Health fiscal year 2003 awarded to Soonmyung Paik
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
S Paik has received honorarium less than $10,000 in total amount during the past two years from Genomic Health, Inc for invited lectures arranged by the company. Genomic Health Inc developed and markets Oncotype DX™ assay as a commercial reference laboratory. Dr Paik is listed as one of the co-inventors in patent filings for Oncotype DX™ assay with all rights released to NSABP Foundation Inc. He does not hold any position or own stocks of the Genomic Health Inc. He does not have any loyalty payment arrangement for his involvement in developing the Oncotype DX™ test.
Glossary
- MKI67
-
Gene that enclodes an antigen identified by monoclonal antibody Ki-67
- AURKA
-
Aurora kinase A, also known as STK15
- BIRC5
-
Baculoviral IAP repeat-containing 5 (Survivin)
- CCNB1
-
Cyclin B1
- MYBL2
-
Gene for v-myb myeloblastosis viral oncogene homolog (avian)-like 2
- GRB7
-
Growth factor receptor-bound protein 7
- ERBB2
-
v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, a neuro/glioblastoma-derived oncogene homolog. It codes for a transmembrane glycoprotein (HER2) that possesses tyrosine kinase activity.
- BCL2
-
B-cell chronic lymphocytic leukemia/lymphoma 2
- SCUBE2
-
Signal peptide, CUB domain, EGF-like 2
- MMP11
-
Matrix metalloproteinase 11 (stromelysin 3)
- CTSL2
-
Cathepsin L2
- GSTM1
-
Glutathione S-transferase M1
- BAG1
-
BCL2-associated athanogene
- NATIONAL SURGICAL ADJUVANT BREAST AND BOWEL PROJECT (NSABP)
-
A clinical trials cooperative group supported by the National Cancer Institute (NCI)
Rights and permissions
About this article
Cite this article
Paik, S., Kim, Cy., Song, Yk. et al. Technology Insight: application of molecular techniques to formalin-fixed paraffin-embedded tissues from breast cancer. Nat Rev Clin Oncol 2, 246–254 (2005). https://doi.org/10.1038/ncponc0171
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/ncponc0171
This article is cited by
-
T cell differentiation protein 2 facilitates cell proliferation by enhancing mTOR-mediated ribosome biogenesis in non-small cell lung cancer
Discover Oncology (2022)
-
Integrative analysis of copy number and gene expression in breast cancer using formalin-fixed paraffin-embedded core biopsy tissue: a feasibility study
BMC Genomics (2017)
-
Multilabel immunofluorescence and antigen reprobing on formalin-fixed paraffin-embedded sections: novel applications for precision pathology diagnosis
Modern Pathology (2016)
-
Reliable PCR quantitation of estrogen, progesterone and ERBB2 receptor mRNA from formalin-fixed, paraffin-embedded tissue is independent of prior macro-dissection
Virchows Archiv (2013)
-
Sample parameters affecting the clinical relevance of RNA biomarkers in translational breast cancer research
Virchows Archiv (2013)