Article Text
Abstract
Aims In everyday practice, epidermal growth factor receptor (EGFR) testing is centralised in referral laboratories that receive paucicellular cytological specimens. Ideally, EGFR testing should be carried out in the centre where the patient is diagnosed such that the most cellular slide can be selected from in-house collected cytological material. However, available techniques are little standardised and difficult to be implemented in settings with little expertise in molecular testing. The Idylla EGFR prototype assay is a rapid and fully automated test which may easily be adopted by a wider number of pathological centres. This study assessed whether an Idylla EGFR prototype assay can be reliably applied to cytological lung cancer specimens.
Methods The limit of detection (LOD) of the Idylla EGFR prototype assay was assessed by cell line dilution studies. A total of 10 ng was directly placed inside an Idylla EGFR prototype assay cartridge. Idylla results were compared with fragment length (exon 19 del) and Taqman assays.
Results The Idylla EGFR prototype assay showed an LOD of 1% mutant allele and yielded valid results in 74/76 (97.3%) samples, detecting all the mutant cases (n=32) identified by standard techniques; in addition, Idylla detected two low abundance EGFR exon 19 deletions and two G719X exon 18 point mutations, not covered by our standard reference method.
Conclusions Idylla EGFR prototype assay is sensitive on extracted DNA and can reliably be applied on cytological samples, enabling implementation of EGFR testing even in less experienced diagnostic units.
- LUNG CANCER
- EGFR
- MOLECULAR PATHOLOGY
- CYTOLOGY
Statistics from Altmetric.com
Introduction
Testing for epidermal growth factor receptor (EGFR) mutations is part of the current standard of care in advanced non-small cell lung cancer.1 The recent guidelines from the College of American Pathologists (CAP), International Association for the Study of Lung Cancer (IASLC) and the Association for Molecular Pathology (AMP) recommend EGFR testing in patients with lung adenocarcinoma, regardless of sex, race, smoking history, histological grade or other clinical risk factors.1 Cytological samples are suitable for EGFR testing, which is crucial, since the large majority of patients present in advanced disease stages.1 ,2
In patients with acute deterioration, EGFR testing results should be available as soon as possible, as administration of EGFR antagonists as second-line agents is less efficient than their use in first-line therapy.1 As a general recommendation, tumour specimens should be sent to testing laboratories within three working days of receiving oncologist's requests.1 However, in a previous survey, we showed the period of time between test request and delivery of the sample is in routine practice nearly double the recommended time.3 Laboratories may use any validated EGFR testing method that is able to detect mutations in cytological specimens with as little as 10% tumour cells.1 As a matter of the fact, the competence needed to validate molecular diagnostic assays is beyond most cytopathology laboratories and, even if there was expertise, a larger number of cases is required to run in-house testing cost-effectively.4 Therefore, the current practice is often external centralised testing. Unfortunately, this is fraught with a higher rate of inadequate samples than in-house testing;5 in fact, the primary cytopathologist is often reluctant ‘to sacrifice’ the morphology of malignant cells for DNA extraction, and the smear sent to centralised laboratories is often paucicellular.6
Thus, ideally, EGFR testing should be carried out in the same centre where the patient is diagnosed. To this end, automated allele-specific real-time PCR (RT-PCR) technology is advancing at rapid pace. In particular, the fully automated molecular diagnostics system Idylla (Biocartis, Mechelen, Belgium) is a fascinating technology.7–9 Sample preparation is combined with PCR thermocycling and fluorescence detection of target sequences. Without needing highly skilled staff, within approximately 90 min, the European Community (CE)-in vitro diagnostic use (IVD) marked Idylla mutational tests can genotype v-Raf murine sarcoma viral oncogene homolog B (BRAF) and Kirsten rat sarcoma viral oncogene homolog (KRAS) with 1% detection limit.7–9 More recently, Biocartis developed an Idylla EGFR test prototype. Although, the Idylla tests were designed for use with formalin-fixed paraffin-embedded (FFPE) sections, the Idylla system can also process DNA preparations from cytological samples, as previously shown.10 The aim of the present study was to assess the Idylla EGFR test performance on lung cancer cytological specimens.
Methods
Study design
Study design is graphically shown in figure 1. Preliminarily, the Idylla EGFR Mutation Assay analytical sensitivity was assessed on DNA derived from cell lines. Then, Idylla EGFR Mutation Assay was carried out on DNA preparations previously extracted from routine cytological samples (n=76) quantified (ng/µL) and tested for clinical reporting by in-house PCR-based methods (fragment length and TaqMan assays) as previously described.11 In cases of discrepancies, next generation sequencing was used as an orthogonal technique, following our previously validated protocol.12 In 17 cases, an additional direct smear was available and Idylla EGFR Mutation Test was also carried out directly on cytological material exploiting the whole Idylla system workflow, as explained below.
Study design: Idylla epidermal growth factor receptor (EGFR) Mutation Assay was carried out on DNA preparations previously extracted from routine cytological samples (n=76) and compared with in-house validated PCR-based methods (fragment length and TaqMan assays), In 17 cases, an additional direct smear was available and Idylla EGFR Mutation Test was also carried out directly on cytological material exploiting the whole Idylla system workflow. NSCLC, non-small cell lung cancer.
The Idylla system
The Idylla system is a molecular diagnostic device for detection of genetic mutations based on automated quantitative allele-specific RT-PCR.7–9 The instrument is composed of a sample preparation module integrated with a combined PCR thermocycling and fluorescence detection module. All consumables required to perform sample preparation and RT-PCR detection are provided in a single disposable cartridge that is loaded into the Idylla system to enable the simultaneous detection of up to 30 molecular targets from a variety of solid and liquid sample types, including DNA preparations from routine cytological specimens.10 Closing of the cartridge after inserting the sample avoids cross-contamination.
The Idylla EGFR Mutation Assay
The Idylla EGFR Mutation Assay is a single-use cartridge-based test. Via microfluidic channels in the cartridge, nucleic acids are transported into five separate PCR chambers, which contain predeposited PCR reagents in dried form (ie, primers, probes, enzymes) designed for the qualitative detection of 18 types of genetic changes for which 53 different mutations have been validated. These include three different point mutations: p.G719S (c. G2155A), p.G719C (c. G2155T), p.G719A (c.G2156C) in exon 18; six different families of deletions ranging from 9 to 24 bp in exon 19; five different insertions (p.InsASV9, p.InsASV11, p.InsSVD, p.InsG, p.InsH) and two point mutations p.S768I (c.G2303T) and p.T790M (c.C2369T) in exon 20 and two point mutations p.L858R (c.T2573G) and p.L861Q (c.T2582A) in exon 21. Detection of these specific targets is performed using fluorescently labelled probes. The analytic time required to perform the results is approximately 2.5 hours, with a hands-on time of less than 2 min.
Analytical sensitivity assessment
The limit of detection of the Idylla EGFR prototype assay was assessed by cell line dilution studies. The mutated PC9 (harbouring EGFR p.E746-A750del) and H1975 (carrying L858R point mutation) and the A549 (EGFR wt) cell lines were serially mixed at dilutions of 25%, 10% and 1%.
Idylla EGFR Mutation Assay on routine cytological specimens
We retrospectively selected from our archive 32 patient DNA samples by the following criteria: (1) the presence of EGFR exon 19 deletion or exon 21 L858R mutation assessed by our laboratory validated assay (our laboratory is registered in the Italian Society of Pathology EGFR external quality control scheme, http://www.egfrquality.it) and (2) the availability of a sufficient (10 ng) amount of stored genomic DNA. Overall, 25 cases harboured an exon 19 deletion, whereas 7 cases carried the L858R point mutation. To ensure specificity of the Idylla EGFR prototype assay, a total of 44 additional EGFR wild-type DNA samples extracted from routine cytological specimens were selected. Overall, a total of 65 cases had ≥20% of neoplastic cells, whereas 11 cases had <20% of cancer cells (table 1). These latter smears had been wholly scraped, since featured a stochastic distribution of malignant and non malignant cells, which precluded tumour component enrichment by manual microdissection. Of these, 59 were Papanicolaou and 17 were Diff-Quik. Archived DNA was processed by the Idylla EGFR Prototype Mutation Test between November 2015 and March 2016. As previously shown, we have directly pipetted 10 ng of extracted genomic DNA in the prototype EGFR cartridge.
Idylla EGFR Mutation Assay performance on archival extracted DNA
A total of 17/76 study cases featured either an air-dried Diff-Quik or a Papanicolaou stained cytological slide, in addition to slides that were previously used for DNA extraction. Any cytological slide was microscopically reviewed, annotating the neoplastic cell percentage and incubated in xylene for 3 days to allow coverslip removing. Each slide was dried and wholly scraped into an Idylla EGFR cartridge.
Analysis of collected fluorescent signals
Conversely to the Idylla BRAF and KRAS mutation tests, where Idylla Console autoanalyses the PCR curve to determine the presence or absence of a mutation and the results are presented on screen as either ‘no mutation detected’ or ‘mutation detected’,7–9 the raw data obtained by the Idylla EGFR prototype on both DNA preparations and scraped cells were centrally analysed at Biocartis; this analysis was blind to the results previously obtained by our standard reference method. In principle, a cycle value (Cq) was calculated by Idylla beta version software for every PCR curve. The presence of a mutant genotype was determined by calculating the difference between the control and sample Cq (ΔCq). The mutant signal is considered valid if the ΔCq is within a validated range. All samples with a valid wild-type signal but a ΔCq value outside the validated range were characterised as EGFR mutation negative. Cases showing invalid results were further investigated to assess the reason behind the failure by evaluating DNA quality by a microfluidic platform based on electrophoretic system 4200 TapeStation (Agilent, Santa Clara, California, USA).
Results
EGFR Idylla prototype sensitivity was evaluated on cell line-derived DNA. As reported in the methods section, we created dilutions down to 1% for both exon 19 and 21 EGFR mutations in wild-type DNA background. In any of the dilutions tested, the presence of EGFR mutant alleles was detected by the EGFR Idylla prototype.
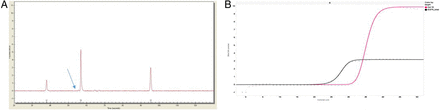
The Idylla results relative to the analysis of 76 cytology samples derived archival DNAs were compared with those obtained by our laboratory validated assays and reported in table 1 (figure 2). Briefly, Idylla yielded valid results in 74/76 (97.3%) samples in a first run. The two invalid samples, showing highly degraded DNA (figure 3), were excluded from the analysis. All 32 mutant cases were confirmed by Idylla, showing a sensitivity of 100%. In addition, in the group of 44 wild-type samples by our standard techniques, Idylla gave a discordant result in four cases. In particular, EGFR Idylla prototype revealed in cases 34 and 40 an EGFR exon 19 deletion, whereas cases 32 and 62 featured a G719X exon 18 point mutation, not covered by our standard reference method. Only these two latter cases could be further investigated by next generation sequencing (NGS), as cases 34 and 40 did not have sufficient residual DNA. In cases 32 and 62, the presence of G719A and G719C mutations was confirmed by NGS analysis.
Discordance between standard reference method and Idylla relative to epidermal growth factor receptor exon 19. Case #40. (A) Fragment length electropherogram showing a clear wild-type allele peak and an additional very low peak of uncertain significance (B), Del15 mutation was detected by using the Idylla assay; note that two curves are detectable, one corresponding to wild-type allele and the other one to Del15-mutated allele.
Archival DNA from a case showing epidermal growth factor receptor Idylla invalid result (#60) further investigated by a microfluidic platform based on electrophoretic system (4200 TapeStation, Agilent) to assess the reason behind the failure. Note that the electrophoretic profile features several peaks due to DNA fragmentation.
In 17/76 cases, an additional air-dried Diff-Quik stained cytological slide was available, and the EGFR Idylla prototype was carried out also by scraping cytological material directly in the cartridge. When the results were compared with those obtained on matched archival DNA, most of the cases (15/17) gave concordant results. Only two cases showed a discordancy, featuring an exon 19 deletion (case n. 15) and an L858R (case n. 25) point mutation detected only by EGFR Idylla prototype on archival DNA. Since Idylla system does, at the moment, not allow the withdrawal of extracted DNA from the cartridge, the discrepancies between archival DNA and directly scraped cytological material could not be further assessed.
Discussion
Our data demonstrate that the Idylla EGFR Mutation Assay is a highly sensitive method that can consistently detect EGFR mutations with as little as 1% mutant DNA in a wild-type background, as shown in cell line studies. Thus, the Idylla EGFR Mutation Assay meets the CAP/IASLC/AMP guideline strong recommendation to use methodologies able to detect EGFR mutations in specimens with only 10% cancer cells.1 This requirement is particularly appropriate when testing cytological samples that often feature a stochastic distribution of malignant and non-malignant cells, which precludes tumour component enrichment by manual microdissection.2 In our series, the EGFR Idylla prototype showed a good performance, enabling in 100% of cases (32/32) the EGFR mutational status confirmation. In addition, this assay was able to resolve cases with low-intensity peaks on the fragment assay electropherograms of the uncertain technical interpretation but suggestive of clinical significance. In particular, cases n. 34 and 40, reported as negative by our reference methods were defined as mutant by EGFR Idylla prototype, as illustrated in figure 2, probably reflecting a higher sensitivity level. In addition, while these latter techniques cover only exon 19 deletions and L858R mutation, EGFR Idylla prototype has a broader reference range, as shown by cases 32 and 62, in which Idylla detected G719X exon 18 point mutations, also confirmed by our validated NGS approach. Noteworthy, the CAP/IASLC/AMP guideline strongly recommends to extend the EGFR mutational assessment also to exons 18 and 20 for the most common mutations1 and for lower prevalence variants, whose response to EGFR antagonists may differ.13
Besides a high analytical sensitivity and a large reference range, the main advantage of the Idylla system is the possibility to extend the feasibility of EGFR testing to smaller laboratories, fully exploiting the automated workflow, which reduces the time required to genotype DNA for EGFR mutational status assessment in approximately 2.5 hours, compensating for the high cost of a single cartridge (∼€200). Until recently, the automated Idylla tests for mutation detection in predictive molecular pathology were limited to histological samples. This reflected the manufacturer design to process FFPE sections. We here showed that the Idylla EGFR prototype test was able to analyse also cytological samples, which is crucial since these specimens represent the most common available material to select patients for targeted treatments in clinical practice. In addition, we also showed that only 10 ng of archival DNA, directly pipetted into the cartridge, is sufficient to obtain results in most samples (74/76). Only two cases showing marked DNA degradation (figure 3) yielded an invalid result. The demonstration that Idylla reliably processes extracted DNA suits cytological practice, where often only a single slide is available. In this study, we also investigated the possibility to directly scrape cytological material from de-coverslipped archival smears into the cartridge; in fact, the sample preparation module of Idylla system uses high-frequency intensity focused ultrasound technology to enable automated DNA extraction. This enables performing mutation testing even without the basic expertise and equipment to perform DNA extraction and purification. While previous studies showed that a full concordance can be obtained between FFPE sections and extracted DNA, in this study, when scraping cytological material directly into the cartridge, cases 15 and 25 gave a false-negative result. Since in both cases the scraped material contained a tumour cell percentage sufficient for EGFR mutational status assessment (table 2), it is conceivable that the original cytological staining might have interfered with the detection of the fluorescence being developed during target amplification. Thus, further technological refinements are needed to better adapt the automated extraction modalities to stained cytological material. However, the possibility to use directly the extracted DNA may overcome the limitations in number of smears and abundance of material, inherent to cytological specimens. Furthermore, aliquoting of the extracted DNA may offer the opportunity to perform additional biomarker testing and to run an orthogonal technique for result confirmation.
Idylla EGFR Mutation Assay performance on scraped cytological samples
In conclusion, we showed that the fully automated molecular diagnostics system Idylla represents a promising option for EGFR mutation testing on cytological samples of lung cancer to enable taking, also in less experienced laboratories, very rapid treatment decisions with high sensitivity, large reference range and ease of use.
Take home messages
Ideally, epidermal growth factor receptor (EGFR) testing should be carried out in the same centre where the patient is diagnosed.
The Idylla tests were designed for use with formalin-fixed paraffin-embedded sections. We showed that it can also process DNA preparations from cytological samples.
Idylla enables, even in less experienced laboratories, EGFR testing in 2 hours, allowing very rapid treatment decisions with high sensitivity, large reference range and ease of use.
References
Supplementary materials
Abstract in Italian
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
- Abstract in Italian - Online abstract
Footnotes
This study was in part presented at the 2016 american society of clinical oncology (ASCO) Annual Meeting.
Handling editor Runjan Chetty
Contributors Conceptualisation: CDL, UM and GT; Methodology: CDL, GG, PP, EV and UM; Writing—Original draft: CdL; Writing—review and editing: CdL and GT; Supervision: GT.
Funding Funding was provided by University Federico II of Napoli. The Idylla system was provided on loan from Biocartis with a sample set of cartridges.
Competing interests None declared.
Patient consent Obtained.
Provenance and peer review Not commissioned; externally peer reviewed.