Article Text
Abstract
The p16 gene belongs to INK4 family of genes and is made up of four members: p16 INK4A , p15 INK4B , p18 INK4C and p19 INK4D , all of which share biological properties, namely, inhibition of cell growth and tumour suppression. After p53, p16 is the second most common tumour suppressor gene. It has been regarded as the familial melanoma gene. Immunohistochemistry for p16 has a well-defined role in distinct pathological scenarios. It is used to distinguish desmoplastic melanoma from reactive fibrous proliferation, with former showing strong nuclear positivity. In other types of melanoma, p16 protein expression is lost. Spitz nevi show retention of nuclear staining for p16. Benign mesothelial proliferations tend to retain nuclear p16 immunoreactivity, while malignant mesotheliomas lose expression. However, p16 fluorescent in-situ hybridisation analysis is recommended in the workup of malignant mesothelioma. Another common application of p16 immunohistochemistry is as an indicator for human papillomavirus (HPV) infection and p16 protein is overexpressed in HPV-associated tumours. In this context, p16 immunopositivity should be strong, diffuse, nuclear or nuclear and cytoplasmic in location. Another use for p16 is demonstration of p16 immunopositivity in well-differentiated and dedifferentiated liposarcoma.
- molecular genetics
- genetics
- histopathology
Statistics from Altmetric.com
p16 is a tumour suppressor gene and goes by several names: MTS-1 (major tumour suppressor 1), INK4a (inhibitor of cyclin-dependent kinase 4a) or p16INK4 and CDKN2A (cyclin-dependent kinases inhibitor 2A). Its location at 9p21 is the site of loss of heterozygosity (LOH) in several malignancies and unsurprisingly, p16 is thus implicated in several tumours1 .
The emphasis of this review will be centred on the lesions and tumours wherep16 and its protein expression are of diagnostic or potentially diagnostic value. Other tumours in which p16 is abrogated will be covered briefly for completeness.
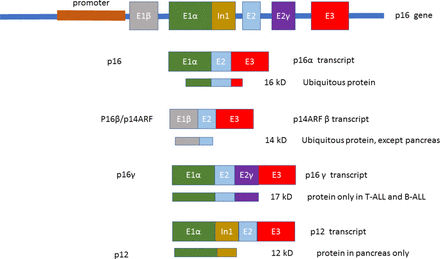
p16 gene
The INK4 family includes four members: p16 INK4A , p15 INK4B , p18 INK4C and p19 INK4D , which show analogous biological characteristics involved in inhibition of cell growth and in tumour suppression.1 2
The tumour suppressor p16 gene encodes proteins involved in the regulation of two fundamental cell cycle pathways, the p53 and the RB1 pathway.
The INK4A locus is localised in short arm of chromosome 9 at position band 21.3(9p21.3).3 Using alternative exons, the p16 gene generates four transcriptional variants: p16INK4A, a cyclin-dependent kinase inhibitor, p14ARF (alternative reading frame), which binds to MDM2,4 p12 and p16γ.5
The structure of the p16 gene includes exons E1β, E1α, E2, E2γ and E3 (figure 1).
Schematic representation of the p16 gene (adapted from Li et al 5).
Exon E1α is located in the 5’ region of the p16 gene. E2γ is an alternatively spliced exon present between exon E1α and E2.6
The alternative splicing of E1α versus E1β to a shared additional exon present in the INK4A gene produces mRNAs that results in E2 sequences with two diverse reading frames.
The transcript with E1α encodes p16INK4a while the transcript with E1β encodes p14ARF using an alternative AUG present in the E1β exon.
Thus, the splicing of exon E1α onto exons E2 and E3 generates the tumour suppressor p16.
The splicing of exon E1β onto exons E2 and E3 generates the tumour suppressor p14ARF. The sequence of exon E1β does not show any similarity with the one of exon E1α, which is transcribed using a different promoter. In addition, the reading frame of p14ARFis shifted by only a base pair; therefore, exons E2 and E3 have a new (ARF) with reference to that of p16. Consequently, p14ARF has a different structure compared with p16 and shows diverse biological activities.5
The p12 transcript has exon E1α in common with p16; using a different splice site present in intron 1, a continuous sequence of 274bp present in intron 1 is transcribed, before splicing onto p16 exons E2 and E3. The p12 transcript seems to be present exclusively in human pancreas.5
p16γ transcript results from the splicing of the cryptic E2γ, present in p16 intron 2, onto p16 exons E2 and E3. High levels of p16γ are detected in a primary T-ALL and a neuroblastoma cell line, while low levels are present in p16-expressing primary T-ALL and B-lineage-ALL samples.5
p16 protein structure
The p16 protein comprises four ankyrin repeats (AR) (figure 2). ARs are fairly preserved motifs of about 31–34 residues and consist of helix-turn-helix conformation. Adjacent ARs are connected together by loops of different length, which are orientated perpendicularly to the helical axes. The 4 AR motifs are arranged in a helix involved in the binding with the target proteins.
Schematic representation of the p16 protein. Each ankyrin repeat consists of a helix-turn-helix (H-T-H) structure. The four H-T-H motifs are connected by three loops in beta and gamma turn structure (adapted from Byeon et al 24).
When CDK6 binds to the cavity of p16, it exposes its catalytic cleft to p16 inducing an electrostatic interaction between D84 of p16 and R31 of CDK6, possibly causing a decrease in the kinase activity.
Cancer-related mutations at residue D84 of p16 eliminated this inhibition causing unrepressed cell proliferation. p16 protein could also prevent the action of CDK4/6 by compromising the interaction with cyclin D. In p16, most of the residues, which interact with CDK4, are positioned in the second and third ARs and in the loop connecting these two ARs.5
p16 function
p16 negatively regulates the pRb-E2F pathway during the cell cycle. In G0 and initial G1, pRb is hypophosphorylated and aggregates with E2F transcription factors, preventing E2F transcription factors to interact with the promoters of genes involved in proliferation (cyclin B1, dihydrofolate reductase, JUNB and others). When the cell proliferates, pRb undergoes increasing phosphorylation by CDK4 and CDK6 at the end of phase G1, driving the progression into S phase. The binding of p16 to CDK4 and CDK6 results in hypophosphorylation of pRb. In addition, p16 can disturb the complexes between CDK6/4 and non-p16 inhibitors, increasing the availability of non-p16 inhibitors, inhibition of CDK2 activity and increase of hypophosphorylated pRb. All these events result in arrest of cell proliferation at the G1/S limit. p16 interacts with TFIIH, an essential transcription factor, preventing the phosphorylation of the C-terminal domain the subunit of RNA polymerase II in the C-terminus, resulting in cell cycle arrest.
p16 also interacts with JUNK1 and JUNK3 suppressing their kinase activity.
It has also been reported that p16 increases abnormally with ageing and that elevated levels of p16 induce senescence of progenitor cells and premalignant tumour cells.5
Genetic alteration
The frequency of p16 inactivation in cancers range from 20% in breast cancer to 85% in pancreatic adenocarcinoma.5
The genetic alterations inducing inactivation of p16 include point mutations, homozygous deletions, promoter hypermethylation and LOH.
The most frequent genetic alterations of p16 are homozygous deletion and promoter hypermethylation.5
It seems that there is a predilection for a particular kind of p16 alteration in certain tumour types: homozygous deletions of p16 are more frequent in pancreatic adenocarcinoma (48%), while point mutations (30%) are recurrent in head and neck squamous cell carcinomas.. In gastric adenocarcinoma, aberrant methylation represents the majority of p16 alterations (34%), with p16 deletions or mutations being infrequent (0%–2%).5
The type of genetic alteration defines p16 functionality: homozygous deletions and promoter hypermethylation, typically, result in non-functional p16, while point mutations, particularly missense mutations, and in-frame small deletions, disrupt p16 structure and activity.5
In cancer, many of the missense mutations identified in p16 protein are mainly situated in the second, third and fourth ARs.5
These missense mutations are classified into four groups based on the mutagenic effect they have on the CDK4-inhibitory capacity, structural stability and protein integrity:
Missense mutations of residues implicated directly in the interaction with CDK4/6 are found on the concave aspect for CDK4 binding, resulting in preserved protein structure, similar conformational stability with prominent reduced CDK4-inhibitory activity.5
Missense mutations that affect residues indirectly implicated in CDK4 binding, affect the stability of p16/CDK4 interactions leading to a sensible reduction of CDK4-inhibitory function, analogous conformational stability with non-significant structural alterations.5
Missense mutations that involve residues implicated in forming the core structure of p16 disrupt the entire structure of p16 and consequently abolish its CDK4-inhibitory function.5
Missense mutations in the first, fourth ARs and the N and C-termini are implicated in binding to different factors such as TFIIH, JNKs and GRIM-19 or are involved in phosphorylation, without causing any obvious modifications in the in the inhibition of CDK4 , stability or structure.5
Regulation of the expression
Regulation of p16
Transcription of p16 shows several ways of control, many of which are regulated in several ways mostly via the regulatory elements located in the p16 promoter.
Ets1 and Ets2 (E26 transformation-specific) transcription factors are targets of Ras-Raf-Mek pathway mediated through phosphorylation by MapKinases. A conserved Ets-binding site is located in the p16 promoter containing a conserved Ets binding site. Ets1/Ets2 are positive regulators of p16 expression.5
The latent membrane protein 1, encoded by the Epstein-Barr virus, suppresses the p16 promoter by endorsing the exporting of Ets2 from the nucleus with inactivation of Ets2-mediated transactivation.5
An E-box (enhancer box) is a DNA response element that acts as a protein-binding site and regulates gene expression. Two E-box elements are present in the p16 promoter.
E47 has a HLH domain which can regulate gene expression. On binding to the 2 E-boxes, E47 activates the expression of p16 in ageing cells.5
On the other hand, the heterodimer E47-Id1 binds to E-box elements, leading to the inhibition of the p16 promoter.5
Myc also binds to the E-boxes present in the promoter region and the first intron of p16, thus upregulating p16.5
The p16 promoter guanine cytosine (GC)-rich region comprises no less than five putative GC boxes for Sp transcription factors, including a positive transcription regulatory element for Sp1 binding.5
The HMG box-containing protein 1 (HBP1) is a transcription factor in the Ras-Raf-Mek signalling pathway, which has a binding site in the p16 promoter also containing a binding site for the HBP1, a transcription factor in the Ras-Raf-MEK pathway and this may play a role in the ageing process mediated by p16.5
In addition, the p16 promoter contains a binding site for theINK4atranscription silence element, a negative regulatory element.5
Peroxisome proliferator-activated receptor alpha (PPARα) inhibits cell cycle progression at the G1/S transition. The binding of PPARα to the peroxisome proliferator response element (PPRE) region and interaction with Sp1 bound to the p16 promoter leads to overexpression of p16 mRNA.5
There is also a complex control of p16, p14ARF and p15 cluster of genes, which involves several mechanisms. PcG proteins, which include Bmi1, Cbx7, Ring1b or Phc2, are transcriptional repressors that modulate histone changes, resulting in the silencing of transcriptional chromatin, particularly that of groups of genes. Expression of PcG proteins leads to downregulation of p16, p14ARF and p15.5
Post-translational regulation
Phosphorylation of p16 is increased in senescence epithelial cells. Phosphorylated p16 displays enhanced binding affinity with CDK4/6 compared with unphosphorylated p16, leading to G1 arrest in senescence. Different residues of p16 may be phosphorylated and phosphorylation may eliminate most of p16 CDK4-inhibitory activity, affecting the core structure or the conformational stability. In addition, any event that affects p16 phosphorylation may pertubate p16 activity.
Protein phosphorylation is also strictly linked to the degree of oxidative stress inside cells. Thus, oxidative stress can promote phosphorylation of p16 phosphorylation, leading to arrest of cell division and early senescence, preventing cells from becoming neoplastic.5
Many proteins positively/negatively regulate the p16/CDK4 interaction and consequent CDK4-mediated phosphorylation of pRb.5
Inhibitor KIP proteins (CDK inhibitor proteins), such as p21, p27 and p57, are able to inhibit the majority of the CDK-cyclin complexes along with some kinases unrelated to CDKs.5 The KIP/CDK4/6 complex may dislocate p16 from CDK4/6 enabling p16 to be involved in other pathways apart from phosphorylation via pRb CDK4/6 or to be degraded.5
GRIM-19 suppression is accomplished by direct interaction with p16, thus promoting p16 inhibition of pRb phosphorylation by CDK4/6. CDK4, p16 and GRIM-19 form a ternary complex. GRIM-19 promotes the binding of p16 to CDK4, while the overexpression of cyclin D1 results in loss of the CDK4/P16/GRIM-19 complex.5
p16 can bind to and suppress NF-κB, IκBα, an inhibitor of NF-κB, which contends with p16 for CDK4 binding and inhibits phosphorylation of pRb by CDK4..5
Gankyrin and p16 compete for binding to CDK4; however, CDK4 is not inhibited by the binding to Gankyrin, thus resulting in cell cycle progression.5
P34SEI-1 binds to only CDK4 which seems to oppose p16 activity, making pRb phosphorylation by CDK4 resilient to p16 inhibition during late G1 phase.5
Tax is a transcription activator encoded by exon 2 of the human T lymphotropic virus 1 genome. This can form a complex with p16 preventing the inhibition of CDK4/6 by p16, resulting in cell cycle progression.5
Regulation of p16 in cancer
Genetic inactivation of p16 is frequently found in cancer.
Diverse mechanisms such as overexpression of cyclin D1 (CCND1), gankyrin (PSMD10), SEI-1 (SERTAD1), CDC6 and NF-κB can affect the regulation of p16 function.
The regulation of P16 in human cancers depends on the effects of the genetic abnormalities of p16, activation of oncogenes and changes in related tumour suppressors.
Partial absence of p16 function secondary to missense mutations of p16 can be balanced by increased p16 levels observed in some tumours.
Even if p16is preserved, additional molecular changes resulting from overexpression of CDC6, cyclin D1, gankyrin and SEI-1 may functionally inactivate p16, causing cancer progression.
In cancers, p16 functionality is not inevitably a reflection of the genetic status of p16 itself. The p16 status secondary to genetic inactivation and oncogene deregulation may differ during different stages of tumour growth: during the earliest stages, deregulation of p16 is most likely secondary to activation of oncogenes and may occur before the genetic alterations of p16.
Finally, oncogene-mediated p16 deregulation may affect p16 activity more dramatically than its genetic inactivation.
The overexpression of p16 in cancers is not well understood. Overexpression of wildtype or mutant p16 is associated with poor outcomes in several malignancies (neuroblastoma, uterine cervix, ovarian, breast, prostate and oesophageal squamous cell carcinomas). Overexpression of p16 mutants may be an attempt to restore p16 functions in some tumours and may be promoted by stress or oncogenic factors but its negative impact on cellular proliferation can be avoided or prevented by alternative molecular events.5
Tumours in which p16 is mutated and use of p16 immunohistochemistry in diagnostic pathology
Malignancies of almost every type have displayed mutations or molecular alterations of p16. Indeed, Liggett and Sidransky suggest that the frequency of p16 inactivation occurs in about 26% of all malignancies, thus making it second to p53 in terms of frequency of cancer involvement.7 Table 1, adapted from their paper, reflects the range of tumours displaying p16 aberrations.7
Cancers and frequency of p16 inactivation (adapted from Liggett and Sidransky7)
The following section highlights the role of p16 immunohistochemistry and/or fluorescent in-situ hybridisation (FISH) in tumours.
Melanoma
p16 is particularly implicated in familial melanomas but is also involved in sporadic cases. Familial melanoma, which accounts for approximately 5%–10% of all melanomas, arises in the setting of familial atypical multiple mole/melanoma (FAMM).7 Germline point mutations in p16 are seen in FAMM kindreds, and because of this, p16 is regarded as the ‘familial melanoma gene’.
A study of the use of immunohistochemistry for p16 in Spitzoid lesions was undertaken.8 Loss of p16 expression was not apparent in Spitz nevi (in other words, p16 immunoreactivity was present/retained), but loss of p16 was seen in 26% of atypical Spitzoid lesions and 16% of Spitzoid melanomas.8 It is interesting to note that desmoplastic melanomas, on the other hand, retain p16 immunohistochemistry and are p16 immunopositive (figure 3). Others have advocated combining p16 immunohistochemistry with Ki-67 and HMB-45 as part of a scoring tool in distinguishing melanoma from benign histological mimicker.9
p16 immunoexpression showing retention (nuclear staining) in a desmoplastic malignant melanoma.
Fatty tumours
The morphological distinction of well-differentiated liposarcoma or atypical lipomatous tumour from benign fat lesions, especially, deep-seated lipomas can be challenging.10 While there are subtle microscopic clues and location is of paramount importance, sometimes one has to resort to ancillary techniques to resolve a diagnostic dilemma. Immunohistochemistry is now part of the armamentarium in the workup of such lesions with p16, CDK4 and MDM2 (including MDM2 amplification via FISH) playing a diagnostic role.11 12 p16 nuclear immunopositivity, in both well-differentiated and dedifferentiated liposarcoma, either alone or in conjunction with CDK4 and MDM2 is of diagnostic help11 12 (figure 4). p16 positivity when coupled with CDK4 positivity is a very sensitive and specific marker for well-differentiated/atypical lipomatous tumour and dedifferentiated liposarcoma.11 12 Immunohistochemistry for p16 in benign fat tumours (lipomas) is negative. An important pitfall to be aware of is nuclear p16 expression in areas of fat necrosis, especially in deep-seated lipomas.13
A well-differentiated (atypical) liposarcoma displaying nuclear p16 immunoreactivity.
Mesothelioma
Chromosome 9p alterations are among the most frequent molecular aberrations seen in malignant mesotheliomas.14 Approximately 30% of malignant mesotheliomas show one of three alterations of the p16 gene: methylation, deletion or a point mutation.14 Immunohistochemistry for p16 shows that almost all benign and reactive mesothelial proliferations retain nuclear positivity for p16. Conversely, malignant mesotheliomas demonstrate loss of nuclear staining for p16. However, it is important to recognise that failure to show loss of expression of nuclear p16 does not exclude a diagnosis of malignant mesothelioma. With this in mind, FISH for p16 has become de rigueur part of the work for malignant mesothelioma. It is a more reliable method of assessing p16 status in mesothelial lesions. In a comparison of immunohistochemistry and FISH for p16, Chiosea and colleagues showed that the former was less sensitive.15 In their series, both benign and malignant mesothelial proliferations showed p16 immunopositivity.15 While benign proliferations more commonly (87% of cases) retained nuclear p16, 60% of malignant lesions also were positive including almost 30% of peritoneal mesotheliomas.15 In view of this, FISH for p16 is recommended. It should also be noted that there is no direct correlation protein expression of p16 and homozygous deletions of the gene. FISH for p16 and BAP1 immunohistochemistry (loss of BAP1 is seen in malignant mesotheliomas) even in effusion cytology specimens is recommended.16
Anogenital lesions
Uterine cervical and anal squamous epithelia are the site of human papillomavirus (HPV) infection and retinoblastoma (Rb) is inactivated by HPV E7. As a result of this inactivation of Rb, there is no longer inhibition of p16 which then accumulates. This is a reproducible enough phenomenon that p16 protein accumulation as detected by immunohistochemistry is regarded as a reliable surrogate marker for infection by high-risk HPV.17
In terms of localisation of p16 protein, it should be nuclear or nuclear and cytoplasmic.18 This was incorporated into the Lower Anogenital Squamous Terminology guidelines that stipulate only diffuse strong nuclear or nuclear and cytoplasmic positivity should be regarded as positive.19 Cytoplasmic only, focal, weak wispy and scattered single cell positivity is regarded as negative.19
Normal squamous epithelium is usually negative for p16 (figure 5A). The diffuse strong nuclear or nuclear and cytoplasmic or ‘block’ positivity covering at least one-third of the epithelial thickness is reflective of high-grade dysplasia (figure 5B). Focal or patchy staining is indicative of a low-grade squamous intraepithelial and maybe associated with low-risk or high-risk HPV genotypes.19
Normal squamous epithelium from the uterine cervix shows absence of p16 immunoexpression (A), while a high-grade dysplastic lesion is diffuse and strongly p16 positive in the nucleus and cytoplasm (B).
Atrophic squamous epithelium, immature squamous metaplasia and transitional metaplasia may all mimic squamous dysplasia and the use of p16 immunohistochemistry helps resolve the diagnostic dilemma as these conditions are usually p16 negative.17 Immature squamous metaplasia can be p16 positive (see below).
Mahajan pointed out some important caveats that need to be borne in mind when interpreting p16 immunostaining.19 One should be aware of p16-negative high-grade dysplastic lesions and correlation with morphology is important; in poorly oriented small biopsies, strong diffuse staining may be hard to evaluate, benign ciliated cells and occasionally immature squamous can display strong p16 positivity and p16 is not useful in separating low-grade squamous intraepithelial lesions from normal and reactive squamous epithelium.13 19
Uterine cervical glandular lesions
Normal uterine endocervical glands are usually p16-negative with weak positivity encountered rarely.17 Similar to squamous lesions of the cervix that are associated with HPV, a similar relationship with HPV exists in glandular lesions; hence, a similar p16 staining pattern is encountered (figure 6). It is only the endocervical adenocarcinomas that are not associated with high-risk HPV genotypes that are p16-negative. p16 immunohistochemistry can be used effectively in excluding adenocarcinoma in situ from benign look-alikes as the former is diffusely and strongly positive and the mimics are negative or only weakly/patchily positive. In this diagnostic scenario, combining p16 immunohistochemistry with MIB1 and bcl2 is advocated.17 20
p16 immunohistochemistry is also useful in separating endocervical adenocarcinoma from endometrioid carcinomas, especially in biopsy or curettage material.17 19 The former shows strong diffuse, positivity while endometrial carcinomas are usually negative or focal positivity. Care must be exhibited in small biopsies where the focal positivity of endometrioid carcinoma may be sampled.
An endocervical glandular carcinoma in situ is strongly p16 positive in a similar fashion to the HPV-driven squamous intraepithelial lesions.
Other gynaecological tumours
Vulval intraepithelial neoplasia associated with HPV is similar to cervical squamous epithelial and is p16 positive.17 p16 is strongly positive in small cell carcinomas, uterine serous and high-grade serous carcinomas of Mullerian origin.19 Leiomyosarcomas are said to overexpress p16 protein and this may help in separating them from smooth muscle tumours of uncertain malignant potential.17
Oropharyngeal squamous carcinoma
Once again, p16 is used as a proxy maker for HPV; p16 immunopositivity reflecting an HPV aetiology to the squamous cell carcinoma (figure 7). However, it has been mentioned that there is false-positive p16 immunoreactivity.21 22 The reason advanced for the discrepancy the lack of reproducibility regarding percentage of tumour positivity, intensity of staining and cellular distribution of the staining.19 It is recommended that 70% of tumour cells show strong, diffuse nuclear or nuclear and cytoplasmic p16 staining for the tumour to be labelled as HPV-associated.23
An oropharyngeal squamous carcinoma (A) showing strong nuclear and cytoplasmic staining in over 70% of the tumour cells (B).
Take home points
p16 is the second most common tumour suppressor gene in all cancers.
p16 is regarded as the familial melanoma gene.
p16 fluorescent in-situ hybridisation analysis is important in the diagnosis of malignant mesothelioma.
p16 immunopositivity is seen in well-differentiated and dedifferentiated liposarcoma.
p16 is a surrogate maker for HPV and is overexpressed in HPV-associated tumours.
p16 immunopositivity should be strong, diffuse, nuclear or nuclear and cytoplasmic in location.
References
Footnotes
Handling editor Cheok Soon Lee.
Contributors Both authors contributed equally.
Funding The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests None declared.
Patient consent Not required.
Provenance and peer review Commissioned; internally peer reviewed.